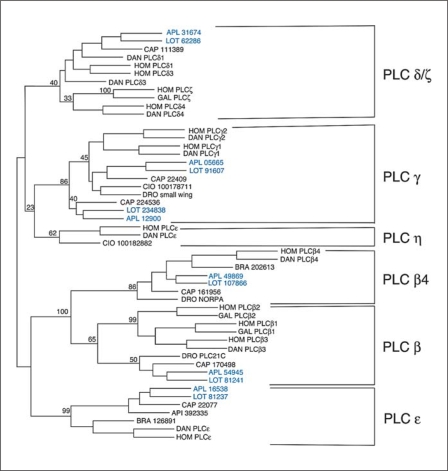
Fig. 2.
Results of a Phylip analysis of merged X and Y domains of PLCs from different species. Six different families were identified, shown on the right. For the vertebrates, human (Hom) and Danio (DAN), the accepted name of the PLC is given. For the primitive chordates, Branchiostoma (BRA) and Ciona (CIO), the protein IDs are given from the NCBI entry. For Apis (API) and Capitella (CAP), the protein IDs are given from the NCBI and JGI genome sites, respectively, and for Drosophila (DRO), the accepted gene names are used. For the mollusks the names are blue. For Aplysia (APL) the contig from the NCBI site containing the X and Y domains is given. For Lottia (LOT) the protein IDs are given from the JGI genome site. The numbers represent the percentage of trees generated by the Phylip protein Neighbour that contained the tree shown. Only selected branches are highlighted to emphasize the division into PLC families. The outgroup (not shown) was yeast (Schizosaccharomyces pombe) PLC.