Abstract
Protein organization on biomembranes and their dynamics are essential for cellular function. It is not clear, however, how protein binding may influence the assembly of underlying lipids or how the membrane structure leads to functional protein organization. Toward this goal, we investigated the effects of annexin a5 binding to biomimetic membranes using fluorescence imaging and correlation spectroscopy. Annexin a5 (anx a5), a peripheral intracellular protein that plays a membrane remodeling role in addition to other functions, binds specifically and tightly to anionic (e.g., phosphatidylserine)-containing membranes in the presence of calcium ion. Our fluorescence microscopy reveals that annexin likely forms assemblies, along with a more dispersed population, upon binding to anionic biomembranes in the presence of calcium ion, which is reflected in its two-component Brownian motion. To investigate the effects of annexin binding on the underlying lipids, we used specific acyl chain-labeled phospholipid analogs, NBD-phosphatidylcholine (NBD-PC) and NBD-phosphatidylserine (NBD-PS). We find that both NBD-labeled lipids cluster under anx a5 assemblies, as compared with when they are found under the dispersed annexin population, and NBD-PS exhibits two-component lateral diffusion under the annexin assemblies. In contrast, NBD-PC diffusion is slower by an order of magnitude under the annexin assemblies in contrast to its diffusion when not localized under anx a5 assemblies. Our results indicate that upon binding to membranes, the peripheral protein annexin organizes the underlying lipids into domains, which may have functional implications in vivo.
INTRODUCTION
The spatial and temporal organization of membrane-associated proteins and lipids is essential for a variety of cellular functions, such as signal transduction, endocytosis, and membrane trafficking (1, 2). Dynamic functional assemblies of proteins and lipids result from protein-protein and protein-lipid interactions (3–6) that are due to van der Waals, steric and electrostatic interactions (7). In addition to these direct interactions, membrane-mediated effects such as hydrophobic mismatch (8) and lipid depletion (9) are also likely to influence protein and lipid organization on membrane surfaces. Although great progress has been made in identifying the key factors in protein-lipid organization, the physical mechanisms responsible for the resulting lateral heterogeneity remain poorly understood due to the dynamic and complex nature of biological membranes.
Our long-term objective is to investigate protein and lipid organization in biomembranes to establish rules that can be universally applied to a set of proteins with similar properties to predict whether proteins will randomly distribute or exist as assemblies or as ramified chains on membrane surfaces. Toward this end, we used annexin a5 (anx a5) as a representative peripheral protein to study its organization on model biomembranes and its binding effects on the lateral diffusion of the underlying lipids. Annexins are a family of peripheral intracellular proteins that bind to phospholipid membranes in a calcium-dependent manner and are widely distributed in a variety of cell types in different plant and animal species (10). In addition to playing functionally important physiological roles in phagocytosis (11) and fibrinolysis (12), annexins are also known to be involved in docking and fusion of exocytotic vesicles with the plasma membranes of secretory cells (13). Annexin mutations have been implicated in a number of human disease states such as antiphospholipid syndrome (14), systematic lupus erythematosus (15), prostate cancer (16, 17) and diabetes (18). Anx a5, a 35 kDa protein which inhibits phospholipid-dependent pro-coagulant reactions in vitro, forms trimers when bound with high affinity to anionic phospholipids such as phosphatidylserine (PS) (19–21).
We hypothesize that for annexin, protein-protein interactions are enhanced as a result of lipid binding, which in turn reorganizes the underlying lipids through extended, weak nonconvalent interactions. To test this hypothesis, we used fluorescence microscopy and fluorescence correlation spectroscopy (FCS) to investigate the distributions and dynamics of membrane-bound anx a5 and several fluorescent lipid analogs in supported planar membranes composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC or 16:0–18:1 PC) in the presence or absence of 40 mol% 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS or 16:0–18:1 PS). We used fluorescence imaging to find that annexin binds to the membrane as assemblies and as a dispersed fraction. Also, we investigated changes in the dynamics of the underlying membrane that were induced by anx a5 binding by measuring the lateral diffusion of both annexin and lipids with FCS (22–24). These protein-induced changes were reflected by distinct differences in the translational diffusion in the presence and absence of anx a5 and depend upon the chemical structure of the lipid analog (e.g., the headgroup or placement of the fluorophore).
RESULTS AND DISCUSSION
Annexin exhibits a high affinity and specificity to POPS-containing membranes in the presence of Ca2+
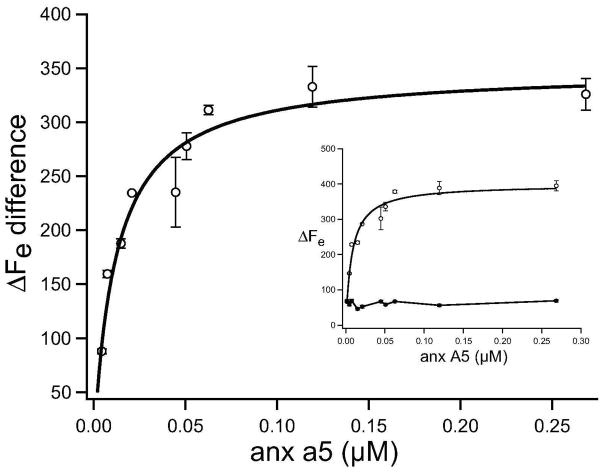
We evaluated the binding affinity of anx a5 with POPS-containing membranes (POPC + 40 mol% POPS) using quartz crystal microbalance (QCM) in the presence and absence of Ca2+. The addition of annexin to these lipid membranes in the presence of 200 μM CaCl2, resulted in an immediate decrease in QCM resonance frequency, indicating protein adsorption on the lipid bilayer. By fitting the binding isotherm to Eq. 3 (Figure 1), a maximum frequency shift (ΔFmax) of 399 ± 15 Hz and a Kd = 8.0 ± 0.5 nM were obtained. These results agree well with the Langmuir model, which assumes a uniform surface with equal anx a5 binding sites and the absence of protein-protein interactions (25). The nanomolar Kd also implies tight binding under our experimental conditions. Similar dissociation constants (0.5–100 nM) for annexin with membranes containing anionic lipids (such as DOPS, di18:1 PS; 1,2-dioleoyl-sn-glycero-3-[phospho-L-serine]) have been previously reported (26). Control experiments showed that protein adsorption did not occur on POPC membranes at any Ca2+ concentration used (0–200 μM) (data not shown). The binding affinity of anx a5 with POPS-containing membranes, in absence of Ca2+, was much weaker (Figure 1, inset) than in presence of Ca2+. To complement the QCM studies, while gaining new insights into the molecular organization, we used fluorescence microscopy to image the organization of membrane-bound annexin on supported membranes.
Figure 1.
Langmuir adsorption isotherm determines the binding affinity of anx a5 to 40 mol% POPS membranes in the presence and absence of Ca2+. The solid line represents the fit to Eq. 3. Individual isotherms are shown in the inset where open and closed squares depict annexin binding in the presence and absence of calcium ion, respectively. Data points are an average of three measurements obtained with three different vesicle preparations and one anx a5 preparation, and the error bars show standard deviation obtained from three different measurements. Kd is 8.0 ± 0.5 nM.
Anx a5 binds to POPS-containing bilayers as protein assemblies with a dispersed fraction
Fluorescently labeled annexin was imaged after it was incubated with the membrane surface. Wild type anx a5 has a single, solvent-accessible cysteine residue, which was labeled with either AlexaFluor 488 C5 maleimide (Alexa488-anx a5) or TexasRed C2 maleimide (TexasRed-anx a5). Before we could use the fluorescently labeled anx a5 for imaging experiments, we ensured that labeling did not affect the binding of annexin with anionic membranes (data not shown) (27). We then incubated the fluorescently labeled anx a5 in the presence and absence of POPS-containing bilayers. Prior to the addition of anx a5, the unlabeled membranes were dark, indicating zero background. When annexin was incubated with 40 mol% POPS bilayers in the presence of calcium (after extensive rinsing to remove unbound or nonspecifically bound protein), we observed a heterogeneous distribution of anx a5 assemblies (that is, the larger clusters in Figure 2a) on a more uniform annexin background (Figure 2a). When we incubated anx a5 with POPS-containing bilayers in the absence of calcium, we observed no fluorescence (data not shown). The fluorescence intensity of anx a5, incubated with POPC bilayers in the presence of calcium, was also negligible.
Figure 2.
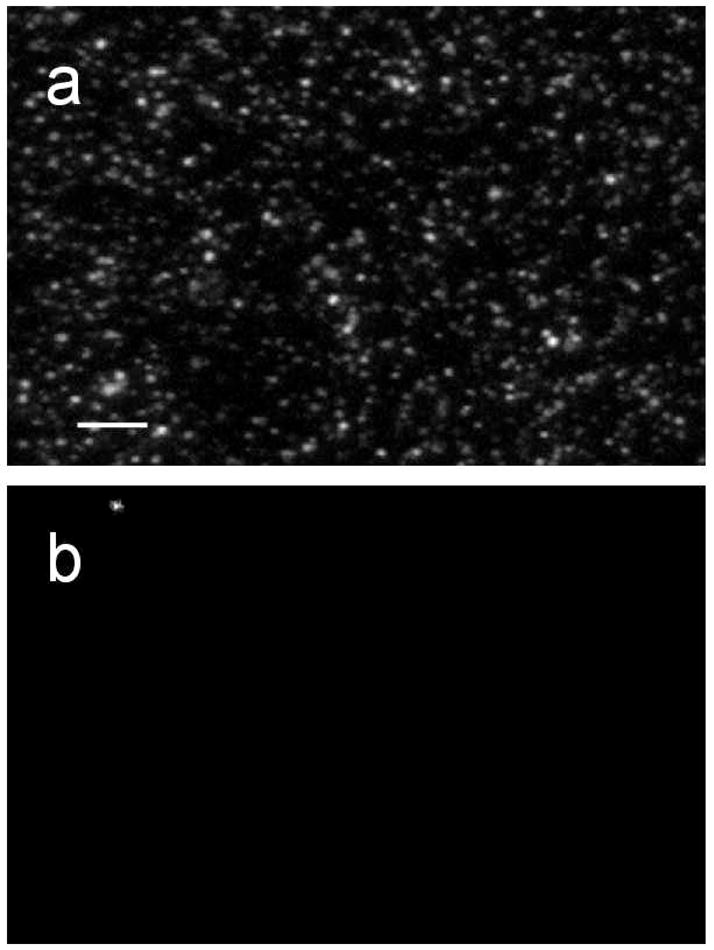
Representative fluorescence images of 0.6 μM Alexa488-anx a5 specifically bound to unlabeled POPC + 40 mol% POPS bilayers (a) and POPC bilayers (b). Bar, 10 μm.
Taken together, these results demonstrate the selective binding of anx a5 to anionic membranes in the presence of Ca2+, which agrees with previous studies in the literature (28–30). Andree et al. (30) used cryoelectron microscopy to investigate the binding of anx a5 to POPS-containing liposomes and observed shape changes in liposomes after annexin binding, which was attributed to the formation of large annexin assemblies that induce surface deformation of the liposomes although their data could not confirm or reject this hypothesis. In a subsequent study, time-dependent growth of two-dimensional monomolecular layers of anx a5 crystals on 20 mol% DOPS in 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, di18:1 PC) supported bilayers was followed using AFM (29, 31). A model describing a two-step process for the two-dimensional array assembly of anx a5 on POPS-containing membranes has been proposed (32). In this two-step model, annexin first binds to several POPS molecules in a Ca2+-dependent manner, which is followed by POPS-bound anx a5 molecules binding to other annexin molecules that are either in solution or membrane-bound. These protein-protein interactions propagate to form two-dimensional arrays of anx a5 assemblies.
As shown in Figure 2a, annexin assemblies were formed on a more dispersed anx a5 background. We attribute the formation of these two-dimensional protein assemblies, as compared to a random protein distribution on the bilayer surface, to the “excluded volume” effect that maximizes entropy (33). In this possible model, the enthalpy of the system is at a minimum when annexin has six annular POPS lipids surrounding it because of the attractive nature of protein and POPS interactions. Approximately three of these POPS molecules are released when an annexin molecule is incorporated into the two-dimensional assembly, resulting in an entropy gain as compared with a random protein distribution, to thermodynamically drive assembly formation on the membrane surface (29). Monte Carlo simulations could, in the future, be used to test this possibility.
Fluctuation autocorrelation of anx a5 reveals two-component diffusion
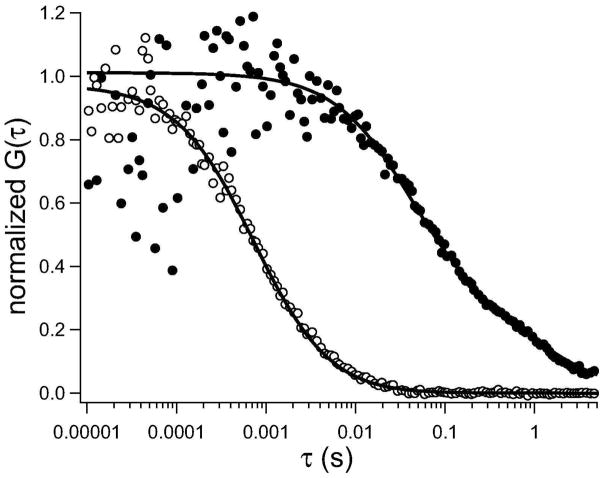
To confirm whether these protein assemblies are stabilized by protein-lipid or protein-protein interactions or both types of interactions, we carried out complementary studies using fluorescence correlation spectroscopy. We examined the translational diffusion of membrane bound-anxa5 when bound to lipid bilayers, as compared with free annexin in solution. The diffusion of Alexa488-anx a5 in aqueous solution is best described by single component diffusion with D = 3.4 × 10−7 cm2 s−1 (n = 34 measurements) (Figure 3, open circles). We further confirmed our initial estimations that annexin is monomeric in solution by calculating the initial amplitude, G(0), from the fits of unnormalized autocorrelation curves (Eq. 4). We can calculate the average number of fluorescent species, N, that are diffusing through the detection volume, following G(0) = N−1 (24, 34). Because we know the concentration of anx a5 in solution (5–200 nM) and the detection volume (1.72 ± 0.51 fL), we calculated N and compared it to the theoretical number of fluorescent molecules in the detection volume (Table 1). If anx a5 is monomeric, the experimental and theoretical values for N will agree, whereas oligomerization would be indicated by the theoretical N being larger than the experimental N. As shown in Table 1, we estimate an oligomerization state of annexin in solution to be 1.26 ± 0.61, which suggest that annexin is primarily monomeric in solution.
Figure 3.
Representative lateral diffusion of annexin in solution and membrane-bound state. Open circles represent the diffusion of Alexa488-anx a5 in solution, and the solid line represents the fit (Eq. 4). Closed circles represent the diffusion of Alexa488-anx a5 specifically bound to POPC bilayers containing 40 mol% POPS in the presence of Ca2+, and the solid line represents the fit to the data points best described by two-component Brownian motion (Eq. 5, m = 2).
Table 1.
Comparison of theoretical and experimental numbers of anx a5 molecules in solution.
| experiment | [anx a5] (μM) | detection volume (fL)a | theoretical N | experimental Na | oligomerization statea (theor N/expt N) |
|---|---|---|---|---|---|
| 1 (n = 18) | 0.20 | 1.4 ± 0.03 | 174 | 174 ± 11 | 1.00 ± 0.02 |
| 2 (n = 22) | 0.10 | 2.5 ± 0.05 | 151 | 139 ± 1 | 1.08 ± 0.01 |
| 3 (n = 20) | 0.02 | 1.5 ± 0.11 | 18.1 | 23.0 ± 0.3 | 0.78 ± 0.02 |
| 4 (n = 20) | 0.005 | 1.5 ± 0.07 | 4.6 | 2.1 ± 0.03 | 2.17 ± 0.04 |
± SD.
The lateral diffusion of membrane-bound annexin was slower than that observed in solution (Figure 3, closed circles), as expected. For Alexa488-anx a5 bound to unlabeled POPS-containing lipid bilayer, the fluctuation autocorrelation curves were fit to two-component, Brownian diffusion, where D1 = (3.1 ± 0.4) × 10−8 cm2 s−1 (f1 = 0.7 ± 0.4; n = 18) and D2 = (1.9 ± 0.7) × 10−10 cm2 s−1 (f2 = 0.3 ± 0.2). The faster diffusion component agrees with typical diffusion coefficients for lipids in biomembranes (see for example Table 2 and 35), and we suggest that it may correspond to the dispersed fraction of annexin trimers (Figure 2a) that may bind to individual or small groups of lipids. The slower component may be due to the larger protein assemblies (observed as clusters in Figure 2a). Our lateral diffusion measurements also agree with single molecule tracking of anx a5 bound to fluid supported bilayers containing 10 mol% PS (36).
Table 2.
Lateral diffusion of chemically distinct fluorescent lipid analogs in the presence or absence of bound anxa5.
| Ca2+? | under anx a5 assemblies? | N (lipid)a | D1a (× 10−8 cm2 s−1) | f1a | D2a (× 10−10 cm2 s−1) | f2a | |
|---|---|---|---|---|---|---|---|
| TexasRed-DPPEb | |||||||
| POPC | |||||||
| + 40 mol% POPS | + | + | 357 ± 23 | 7.3 ± 0.2c | 0.64 ± 0.13 | 3.2 ± 1.1 | 0.36 ± 0.04 |
| + anxa5 (n = 40) | |||||||
| POPC | |||||||
| + 40 mol% POPS | + | − | 250 ± 22 | 3.4 ± 1.5 | 1.0 | — | — |
| + anxa5 (n = 40) | |||||||
| POPC | |||||||
| + 40 mol% POPS | + | NA | 234 ± 43 | 3.4 ± 1.6 | 1.0 | — | — |
| − anxa5 (n = 30) | |||||||
| POPC | |||||||
| + 40 mol% POPS | − | NA | 297 ± 19 | 3.5 ± 1.4 | 1.0 | — | — |
| − anxa5 (n = 30) | |||||||
| NBD-PCb | |||||||
| POPC | |||||||
| + 40 mol% POPS | + | + | 142 ± 17 | 0.74 ± 0.23c | 1.0 | — | — |
| + anxa5 (n = 54) | |||||||
| POPC | |||||||
| + 40 mol% POPS | + | − | 36.3 ± 13.6 | 7.3 ± 1.4 | 1.0 | — | — |
| + anxa5 (n = 21) | |||||||
| POPC | |||||||
| + 40 mol% POPS | − | NA | 32.8 ± 11.2 | 6.3 ± 2.4 | 1.0 | — | — |
| + anxa5 (n = 17) | |||||||
| POPC | |||||||
| + 40 mol% POPS | + | NA | 39.2 ± 27.2 | 5.7 ± 1.3 | 1.0 | — | — |
| − anxa5 (n = 13) | |||||||
| POPC | |||||||
| + 40 mol% POPS | − | NA | 29. 8 ± 5.9 | 6.5 ± 2.4 | 1.0 | — | — |
| − anxa5 (n = 15) | |||||||
| NBD-PSb | |||||||
| POPC | |||||||
| + 40 mol% POPS | + | + | 112 ± 20 | 3.2 ± 1.2 | 0.80 ± 0.22 | 6.8 ± 2.0 | 0.20 ± 0.07 |
| + anxa5 (n = 40) | |||||||
| POPC | |||||||
| + 40 mol% POPS | + | − | 76.6 ± 9.2 | 4.1 ± 1.7 | 1.0 | — | — |
| + anxa5 (n = 40) | |||||||
| POPC | |||||||
| + 40 mol% POPS | − | NA | 90.7 ± 5.2 | 8.1 ± 3.2c | 1.0 | — | — |
| + anxa5 (n = 41) | |||||||
| POPC | |||||||
| + 40 mol% POPS | + | NA | 65.9 ± 9.7 | 4.0 ± 0.2 | 1.0 | — | — |
| − anxa5 (n = 34) | |||||||
| POPC | |||||||
| + 40 mol% POPS | − | NA | 66.2 ± 5.6 | 3.5 ± 0.2 | 1.0 | — | — |
| − anxa5 (n = 31) | |||||||
± SD.
0.1 mol% of TexasRed-DPPE, NBD-PC or NBD-PS was used.
F > Fcrit The fitted data are statistically different from the rest of the cases at 95% confidence limit as assessed by single factor ANOVA. Results were confirmed with the Dunnett’s multiple comparison test.
Anx a5 binding differentially affects the lateral diffusion of lipids in POPS-containing membranes
To investigate the effects of annexin binding on the underlying lipids, we measured the lateral diffusion of chemically distinct fluorescent lipid analogs on and off of anx a5 assemblies, as imaged with fluorescence microscopy. We used the headgroup-labeled lipid TexasRed 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (TexasRed-DPPE) (in conjunction with Alexa488-anx a5), and the acyl chain-labeled probes 1-oleoyl-2-[12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl]-sn-glycero-3-phosphocholine (NBD-PC) and 1-oleyl-2-[12-[(7-nitro-2-1,3-benzoxadiazol-4-yl) amino]dodecanoyl]-sn-glycero-3-phospho-L-serine (NBD-PS) (in combination with TexasRed-anx a5). NBD-PC and NBD-PS are tracers for the diffusion of POPC and POPS, respectively. FCS was used to measure the lateral diffusion in the presence (on and off of annexin assemblies, as visualized by the annexin fluorescence) or absence of anx a5, and in the presence or absence of calcium. For these experiments, annexin was visualized and the laser for FCS was strategically positioned on an obvious annexin assembly (or not). Also note that the supported membranes were composed of 40 mol% POPS in POPC. If any of these lipids co-cluster with annexin, one would expect the lipid to undergo similar diffusive behavior as the protein.
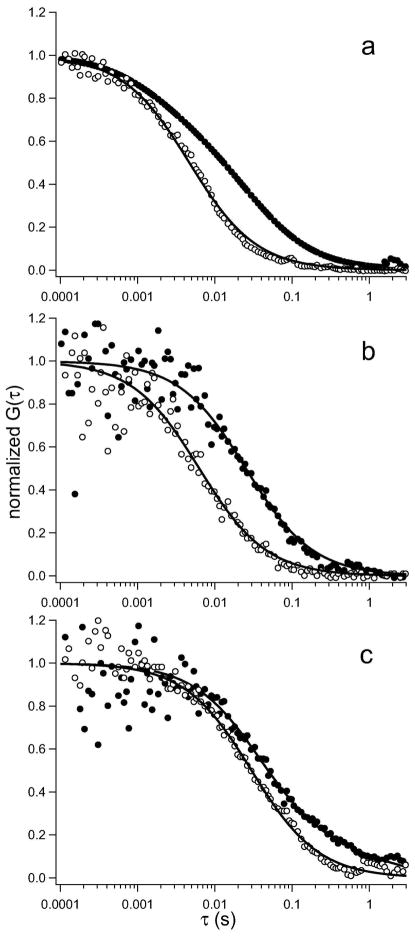
TexasRed-DPPE was used as a probe for general lipid diffusion. The fluorophore is located at the membrane surface and is thus accessible to the protein and solvent. In the absence of bound annexin, TexasRed-DPPE exhibits single component Brownian diffusion with D = (3.4 ± 1.5) × 10−8 cm2 s−1 (n = 30; +Ca2+; Table 2) and D = (3.5 ± 1.4) × 10−8 cm2 s−1 (n = 30; −Ca2+; Table 2). Autocorrelation curves were best described by two-component Brownian diffusion when TexasRed-DPPE is measured under anx a5 assemblies (Figure 4a, closed circles; Table 2). The magnitude of both the faster (D1 = [7.3 ± 0.2] × 10−8 cm2 s−1; f1 = 0.64; n = 40) and the slower diffusion coefficient and (D2 = [3.2 ± 1.1] × 10−10 cm2 s−1; f2 = 0.36; n = 40) agree with the diffusion obtained for labeled annexin using FCS (see above). This agreement suggests a correlation between the protein and TexasRed-DPPE diffusion under the annexin assemblies. When the diffusion of TexasRed-DPPE was measured under the dispersed annexin population, it remained single component, similar to the control experiments in the absence of annexin binding (Figure 4a, open circles; Table 2). The average number of TexasRed-DPPE (N) in the detection volume was calculated from the initial amplitude, as described above. Interestingly, there is a substantial increase in N (~43%) of TexasRed-DPPE measured under the anx a5 assemblies as compared with the protein-free samples or when its diffusion is measured under the dispersed annexin population (Table 2), which may indicate lipid recruitment or confinement under the annexin assemblies. Although these data suggest that annexin binding to bilayers introduces a second slower component to the diffusion of TexasRed-DPPE under the protein assemblies, it is possible that the decrease in diffusion may originate from nonspecific interactions between the anx a5 and the TexasRed headgroup at the water-lipid interface. To assess this possibility, we used acyl chain-labeled fluorescent analogs that also had functional (that is, solvent-accessible) headgroups.
Figure 4.
Representative diffusion measurements of chemically distinct lipid analogs in presence and absence of bound annexin. (a) Alexa488-anx a5 is bound specifically to TexasRed-DPPE labeled POPS-containing membranes in the presence of calcium. (b,c) TexasRed-anx a5 is bound specifically in the presence of calcium to POPS-containing membranes that were labeled with NBD-PC (b) and NBD-PS (c). For all panels, the closed circles represent lipid analog diffusion on annexin assemblies, and the open circles represent diffusion off of anx a5 assemblies. In (a) and (c), the FCS data from the on-assembly experiments were best described by two-component Brownian diffusion (Eq. 5, m = 2; Table 2), and the data obtained off of anx a5 assemblies exhibits single component Brownian diffusion (Eq. 5, m = 1; Table 2). In (b) the FCS data from both on and off the annexin assemblies were best described by single component Brownian diffusion (Eq. 5, m = 1; Table 2).
NBD-PC was used as a probe for the lateral diffusion of POPC. The autocorrelation function of NBD-PC indicates a single diffusing component with D ≈ (6–7) × 10−8 cm2 s−1 (Table 2) away from annexin assemblies and independent of calcium. In contrast, however, when the diffusion of NBD-PC is measured under anx a5 assemblies, a slowly diffusing component was measured with D = (7.4 ± 0.2) × 10−9 cm2 s−1 (n = 54), coupled with a ~75% increase in N when compared to the protein-free sample and when not localized under annexin assemblies (Figure 4b; Table 2). That is, NBD-PC becomes more clustered under the annexin assemblies. These results suggest that annexin binding induces a change in the dynamic structure of lipid bilayer, which in turn affects the diffusion of the zwitterionic POPC.
We then examined the effect of annexin binding on the lateral diffusion of NBD-PS. No significant lipid phase separation was observed upon anx a5 binding (data not shown). Similar to the diffusion of anx a5 and TexasRed-DPPE, we observed two-component Brownian diffusion when NBD-PS is measured under the protein assemblies, with D1 = (3.2 ± 1.2) × 10−8 cm2 s−1 (f1 = 0.80; n = 40) and D2 = (6.8 ± 5.0) × 10−10 cm2 s−1 (f2 = 0.20; n = 40) (Figure 4c; Table 2). Further, NBD-PS is somewhat enriched or clustered (~46%) under these conditions as compared to when it is measured off of protein assemblies or the absence of annexin or in the presence or absence of calcium (Table 2). The diffusion of NBD-PS in these control samples exhibited single component diffusion (D ≈ [3–8] × 10−8 cm2 s−1; Table 2). The two-component Brownian diffusion can be interpreted in a number of ways. First, anx a5 induces two phases of differing composition in POPS-containing bilayers and NBD-PS partitions into both phases. This effect was observed for anx A4 when fluorescence photobleaching recovery was used on 76.5 mol% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG or 16:0–18:1 PG) in POPC membranes (37). We do not observe obvious phase separation (data not shown), although we cannot rule out the possibility of protein-induced nanoscopic domains that are relatively enriched in POPS. Second, anx a5 acts as diffusion obstacles that would be detected as anomalous diffusion (38, 39), which we do not observe. Third, which is the more likely scenario, anx a5 clusters with POPS lipids to form proteolipidic complexes, which lead to decreased diffusion because of the larger-sized assemblies diffusing as units. The similarity in the diffusion behavior of labeled anx a5 and NBD-PS in terms of magnitude of the fast and slow components, suggests the possibility of annexin-POPS complexes stabilized by protein-protein interactions and calcium. We hypothesize that the fast component corresponds to dispersed annexin bound to lipid or lipid not associated with the protein. Interestingly, NBD-PC diffusion is also affected by annexin assemblies. The substantial reduction in the diffusion may be due to NBD-PC becoming hindered as a result of the existence of anx a5–POPS complexes in the bilayer. We should also note that the polar NBD group on C12-labeled lipids has been found to prefer the aqueous interface rather than the hydrophobic interior of the bilayer (40–43) and this may hinder the fluorescent analogs from acting as true mimics of PC and PS. However, we have found that the low amounts of NBD lipids (0.1 mol%) used in this study do not affect annexin binding to the bilayer, as assessed by isothermal titration calorimetry (K. Knutson and A. Hinderliter, unpublished results). Regardless, the distinct differences we observe for NBD-PS and NBD-PC underpin changes in membrane organization that occur upon annexin binding.
CONCLUSIONS
Our microscopy and FCS studies suggest that membrane binding induces protein-protein interactions that lead to annexin assemblies, in addition to a more dispersed population. FCS experiments clearly demonstrate that the binding of anx a5 to PS-containing membranes strongly affects the lateral motion of both POPC and POPS molecules in a lipid specific manner. We hypothesize that upon binding to the membrane, annexin forms a proteolipidic complex that is stabilized by interactions with POPS molecules. This hypothesis is based on the similarity of the PS-bound anx a5 and NBD-PS in PS-containing bilayers bound to anx a5. In these protein-lipid complexes, the POPS lipids experience an environment that differs from that of protein-free lipid bilayers. The proteolipidic complex, in turn, organizes the membrane on the nanoscale to reduce the diffusion of the surrounding POPC molecules.
METHODS
Materials
POPC, POPS, NBD-PS and NBD-PC were purchased from Avanti Polar Lipids. AlexaFluor 488 C5 maleimide, TexasRed C2 maleimide, TexasRed-DPPE, rhodamine green, rhodamine 6G chloride and dithiothreitol were purchased from Invitrogen. Lipids and fluorescent analogs were used without additional purification. Ultrapure water (with a resistivity of >18 MΩ) was used for all buffers used in this study.
Isolation and purification of anx a5
The anx a5 clone (between BamHI and NcoI sites in the pET3d vector) was transformed into chemically competent E. coli BL21 (DE3) cells. Recombinant anx a5 was purified by refolding from the inclusion bodies as described in Elegbede et al. (44) and was ≥95% pure based on densitometry. The wild type anx a5 has only one solvent accessible cysteine (Cys 314), which was used for fluorescent labeling using thiol-reactive probes.
Fluorescent labeling of anx a5
A 40 μM solution of anx a5 in 20 mM HEPES, 100 mM KCl, pH 7.4, was reacted with a 10-fold molar excess of dithiothreitol for 1 h at room temperature. Following reduction, excess dithiothreitol was removed from the protein solution through dialysis using a 12,000–14,000 MWCO membrane (Spectrum Laboratories) against 20 mM HEPES, 100 mM KCl, pH 7.4 (HEPES buffer). Following dialysis, the protein was reacted with a 20-fold molar excess of thiol-reactive dye (either AlexaFluor 488 C5 maleimide [Alexa488-anx a5] or TexasRed C2 maleimide [TexasRed-anx a5] in dimethylsulfoxide) for 24 h at 4°C. Excess dye was removed from the dye-protein conjugate via extensive dialysis against HEPES buffer. The dye/protein ratio was determined using UV-visible spectrophotometry, with a typical dye/protein ratio of 0.7. Protein folding before and after labeling was evaluated by exciting the protein solution at 283 nm and recording the emission spectra using a Luminescence Spectrometer (PerkinElmer, LS 55). An emission maximum at 320 nm confirmed that the protein was not denatured (27).
Small unilamellar vesicle preparation
The day prior to the preparation of the supported bilayers, small unilamellar vesicles (SUVs) of a desired composition (e.g., POPC ± 40 mol% POPS) were prepared as described (45, 46). The top quarter of the supernatant was collected and stored overnight at room temperature and used within 24 h. For some experiments, 0.1 mol% of fluorescent lipid analog (e.g., TexasRed-DPPE, NBD-PC or NBD-PS) was mixed with the lipids prior to SUV preparation.
QCM measurements
A 27-MHz QCM (Affinix Q, Intium Inc.) was used to determine the binding affinity of annexin for lipids in a bilayer. Silica-coated QCM sensors were washed with 200 μL of 1% (w/v) sodium dodecyl sulfate solution, followed by rinsing with water and drying with nitrogen. The surface was then washed twice with freshly prepared piranha solution (3:1 (v/v) concentrated H2SO4 and H2O2) for 5 min, followed by extensive rinsing with water. Cleaned substrates were incubated with 5 μL of SUVs of the desired lipid composition for 30 min to allow complete bilayer coverage, and subsequently rinsed extensively with 50 mM Tris, 100 mM NaCl, pH 7.4 (TBS) to remove unfused vesicles. TBS was then exchanged with 2 mM MOPS, 100 mM KCl, 200 μM CaCl2, pH 7.4 (MBS+Ca). Finally, the sensor cell was filled with 500 μL of buffer, placed in the cell holder and stirred slowly using a magnetic stirrer at 25°C. Lipid bilayer deposition on the silica-coated sensor (91%) was confirmed by measuring the decrease in oscillating frequency of the sensor as SUVs fuse to form bilayer on the silica surface assessed by using the Sauerbrey equation (47),
| (1) |
The frequency change (ΔF) is used to calculate the mass change on the sensor surface (Δb). F0 is the fundamental frequency of the quartz crystal (27 MHz), A is the electrode area (0.049 cm2) (48), ρq is the density of quartz (2.65 g cm−3) (49) and μq is the shear modulus of quartz (2.95 × 1011 dynes cm−2) (50).
To determine lipid-protein affinity, we measured the decrease in oscillating resonance frequency as a function of annexin concentration. As a control, the binding affinity of anx a5 in the absence of calcium was also measured. The decrease in frequency is proportional to number of surface-bound molecules. For the quantitative analysis of the protein binding kinetics, we assumed that the rate-limiting step is the adsorption of protein on the surface and that all binding sites are independent of each other (51). Rate limiting kinetics can be expressed as
| (2) |
where ΔFe is the equilibrium frequency shift for a given protein concentration in solution (canxA 5), and ks is the protein concentration-dependent rate constant (51).
We used Eq. 1, which assumes that the frequency shift is proportional to the adsorbed mass, to obtain the adsorption isotherm by plotting the fitted ΔFe as a function of annexin concentration. The data were fit to obtain the dissociation constant, Kd, and the frequency shift at maximum surface coverage, ΔFmax, following
| (3) |
To determine lipid-protein affinity, differing concentrations of anx a5 diluted in MBS+Ca were added to the bilayer-containing sensor cells and the decrease in oscillating resonating frequency was recorded. As a control, the binding affinity of anx a5 in the absence of calcium (2 mM MOPS, 100 mM KCl, 10 mM EGTA, pH 7.4; MBS–Ca) was also measured. The decrease in frequency is proportional to the mass of molecules adsorbed on the surface.
Supported lipid bilayers and anx a5 binding
SUVs were used to form supported planar bilayers within 24 h of preparation. On the day of an experiment, 75 μL of the SUV suspension was applied to a sandwich made of a detergent-cleaned 3 in × 1 in glass slide and a 22 mm × 22 mm glass coverslip that had been cleaned in argon plasma immediately prior to applying the SUV suspension. SUVs spontaneously fuse to form uniform bilayers. After a 30 min incubation in a humidified chamber, samples were rinsed with TBS to remove unfused vesicles. TBS was later exchanged with either MBS+Ca or MBS−Ca, depending on the particular experiment. The planar bilayers were incubated with 0.6 μM unlabeled or fluorescently labeled annexin (diluted ≥12 h prior to the experiment in either MBS+Ca or MBS−Ca and stored on ice at 4°C until use) in MBS+Ca or MBS−Ca for 15 min at room temperature, and later rinsed with the same buffer to remove any unbound protein. Samples were sealed with VALAP (Vaseline:lanolin:paraffin [2:1:1, w/w]) and imaging or FCS measurements were carried out immediately. Controls for binding specificity and background included bilayers rinsed with MBS+Ca in the absence of annexin, and bilayers rinsed with MBS–Ca in the presence or absence of anx a5.
Fluorescence imaging
Samples were imaged with a Photometrics CoolSnap HQ CCD detector on a Nikon TE2000U inverted microscope with a 60× 1.2 NA objective (Nikon PlanApo) at room temperature (25 ± 1°C). For Alexa488-anx a5, NBD-PS and NBD-PC excitation, a 485/15 excitation filter, 520/20 emission filter and 505 DRLP dichroic were used; and for TexasRed-DPPE excitation, a 555/10 excitation filter, 600/20 emission filter and 560 DRLP dichroic were used. Excitation and emission filter wheels (Ludl Electronic Products) and image acquisition were driven by ISee Imaging software on a Linux-based Pentium class PC (45). Samples were illuminated with mercury lamp excitation, and exposure times were kept constant for a given day of experiments. All images were background and flatfield corrected.
Fluorescence correlation spectroscopy
Confocal FCS was used to characterize the lateral mobility of TexasRed-DPPE, NBD-PC or NBD-PS within supported planar membranes, in the presence or absence of annexin and calcium. For some experiments, the lateral diffusion of Alexa488-anx a5 specifically bound to supported bilayers or in free solution was also measured using FCS. We cannot image a region of interest at the fluorophore concentrations required for an FCS experiment due to the very low signal-to-noise at the fluorophore concentrations (~nM) necessary for obtaining sufficiently large fluctuations that allow correlation (23, 24, 46). Previous to an FCS measurement and after strategically positioning the laser on or off of annexin assemblies as appropriate, we reduced the fluorophore concentration via photobleaching with the mercury arc lamp to obtain an intensity equivalent to 1 × 10−6 –1 × 10−5 mol% as described previously (46).
FCS instrumentation and analysis are described in more detail in Kyoung et al (45). FCS experiments were carried out using either the 488 nm line from a Coherent Innova 90C6 argon ion laser for the bodipy PC or NBD-labeled lipids and Alex488-anx a5 or a 543 nm HeNe laser (Meredith Instruments) for TexasRed-DPPE and TexasRed-anx a5. A focused laser spot was introduced through the epi-port of the microscope and projected onto the sample by overfilling the back focal plane of a Nikon PlanApo 60× 1.2 NA objective. Typical excitation powers ranged from 12–18 μW at the sample plane. A 50 μm diameter optical fiber (OZ Optics) was placed immediately in front of a GaAsP photomultiplier tube (Hamamatsu H7421-40) in a plane conjugate to the sample to limit the detection volume. Correlation curves were acquired with a USB correlator (Flex02-12D correlator, correlator.com) or, in a few experiments, with a PCI bus correlator board (M9003, Hamamatsu) in a Pentium class Windows XP PC.
Data were fit to three-dimensional diffusion with Igor Pro (WaveMetrics) according to (45, 52)
| (4) |
where τ is the time interval, τD is the characteristic diffusion time, and N is the average number of molecules in the three-dimensional Gaussian volume element. G(τ) is the autocorrelation function for three dimensions. We used 1 nM rhodamine green (Drhod green = 2.8 × 10−6 cm2 s−1) (53) or 1 nM rhodamine 6G (DRh6G = 2.8 × 10−6 cm2 s−1) (52) to determine the axial-to-lateral dimension ratio, the structure parameter ωo, for 488 nm and 543 nm excitation, respectively. For our experimental setup, ωo ~ 6–7 was obtained.
For supported bilayer samples, in which either the lipid or protein was fluorescently labeled, we fit data to single and two component diffusion in two dimensions with Igor Pro according to (46, 54, 55)
| (5) |
where G(τ) is the autocorrelation function for two-dimensional diffusion, τ is the time interval, and τDi is the characteristic diffusion time for each fraction (fi, ). For single component, two-dimensional diffusion, m = 1, while for two component diffusion, m = 2 (45, 46, 54, 55). For fluorescence fluctuations due to anomalous diffusion in two dimensions, the correlation function, G(τ) can be modified such that the diffusion time of a molecule, τD, can be calculated following (56)
| (6) |
where α is the anomalous diffusion exponent that is less than unity (56, 57). For all cases, χ2 was used to determine which model best described the data. Diffusion coefficients were calculated following D = ωxy2/4τD, where ωxy is the lateral radius of the detection volume, which is the fiber diameter divided by the objective magnification.
Acknowledgments
We thank A.A. Heikal (University of Minnesota, Duluth, Chemistry & Biochemistry), A.M. Davey and K.M. Krise (Penn State University) for helpful comments on the manuscript, and J. Zhu from correlator.com for his troubleshooting assistance. This work was supported, in part, by the Penn State Materials Research Institute, the Penn State MRSEC under NSF grant DMR 0213623, and the Center for Optical Technologies, which is supported by the Commonwealth of Pennsylvania. Additional acknowledgment is made to the National Institutes of Health grants AG030949 (EDS) and GM64443 (AH), National Science Foundation grant MCB 0718741 (EDS), and Avanti Polar Lipids, Inc., for partial support of this research. We also acknowledge an University of Minnesota Grant-in-Aid for providing partial graduate support (KK).
References
- 1.Andersen OS, Koeppe RE. Bilayer thickness and membrane protein function: an energetic perspective. Annu Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 2.Sachs JN, Engelman DM. Introduction to the membrane protein reviews: the interplay of structure, dynamics, and environment in membrane protein function. Annu Rev Biochem. 2006;75:707–712. doi: 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- 3.Harder T. Formation of functional cell membrane domains: the interplay of lipid- and protein-mediated interactions. Philos Trans R Soc Lond B Biol Sci. 2003;358:863–868. doi: 10.1098/rstb.2003.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poveda JA, Fernández AM, Encinar JA, González-Ros JM. Protein-promoted membrane domains. Biochim Biophys Acta. 2008;1778:1583–1590. doi: 10.1016/j.bbamem.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 5.Liang Q, Ma YQ. Organization of membrane-associated proteins in lipid bilayers. Eur Phys J E Soft Matter. 2008;25:129–138. doi: 10.1140/epje/i2007-10272-6. [DOI] [PubMed] [Google Scholar]
- 6.Marguet D, Lenne PF, Rigneault H, He H-T. Dynamics in the plasma membrane: how to combine fluidity and order. EMBO J. 2006;25:3446–3457. doi: 10.1038/sj.emboj.7601204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohinc K, Kralj-Iglic V, May S. Interaction between two cylindrical inclusions in a symmetric lipid bilayer. J Chem Phys. 2003;119:7435–7444. [Google Scholar]
- 8.Bruinsma R, PIncus P. Protein aggregation in membranes. Curr Opin Sol State Mater Sci. 1996;1:401–406. [Google Scholar]
- 9.Sintes T, Baumgärtner A. Protein attraction in membranes induced by lipid fluctuations. Biophys J. 1997;73:2251–2259. doi: 10.1016/S0006-3495(97)78257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortimer JC, Laohavisit A, Macpherson N, Webb A, Brownlee C, Battey NH, Davies JM. Annexins: multifunctional components of growth and adaptation. J Exp Bot. 2008;59:533–544. doi: 10.1093/jxb/erm344. [DOI] [PubMed] [Google Scholar]
- 11.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 12.Cesarman GM, Guevarra CA, Hajjar KA. An endothelial cell receptor for plasminogen/tissue plasminogen activator (t-PA). II Annexin II-mediated enhancement of t-PA-dependent plasminogen activation. J Biol Chem. 1994;269:21198–21203. [PubMed] [Google Scholar]
- 13.Rescher U, Gerke V. Annexins—unique membrane binding proteins with diverse functions. J Cell Sci. 2004;117:2631–2639. doi: 10.1242/jcs.01245. [DOI] [PubMed] [Google Scholar]
- 14.Rand JH, Wu XX, Andree HA, Ross JB, Rusinova E, Gascon-Lema MG, Calandri C, Harpel PC. Antiphospholipid antibodies accelerate plasma coagulation by inhibiting annexin-V binding to phospholipids: a “lupus procoagulant” phenomenon. Blood. 1998;92:1652–1660. [PubMed] [Google Scholar]
- 15.Cederholm A, Frostegård J. Annexin A5 in cardiovascular disease and systemic lupus erythematosus. Immunobiology. 2005;210:761–768. doi: 10.1016/j.imbio.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Gilmore WS, Olwill S, McGlynn H, Alexander HD. Annexin A2 expression during cellular differentiation in myeloid cell lines. Biochem Soc Trans. 2004;32:1122–1123. doi: 10.1042/BST0321122. [DOI] [PubMed] [Google Scholar]
- 17.Wang LL. Biology of osteogenic sarcoma. Cancer J. 2005;11:294–305. doi: 10.1097/00130404-200507000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Hayes MJ, Moss SE. Annexins and disease. Biochem Biophys Res Commun. 2004;322:1166–1170. doi: 10.1016/j.bbrc.2004.07.124. [DOI] [PubMed] [Google Scholar]
- 19.Oling F, Bergsma-Schutter W, Brisson A. Trimers, dimers of trimers, and trimers of trimers are common building blocks of annexin A5 two-dimensional crystals. J Struct Biol. 2001;133:55–63. doi: 10.1006/jsbi.2000.4337. [DOI] [PubMed] [Google Scholar]
- 20.Funakoshi T, Hendrickson LE, McMullen BA, Fujikawa K. Primary structure of human placental anticoagulant protein. Biochemistry. 1987;26:8087–8092. doi: 10.1021/bi00399a011. [DOI] [PubMed] [Google Scholar]
- 21.Reutelingsperger CP, Hornstra G, Hemker HC. Isolation and partial purification of a novel anticoagulant from arteries of human umbilical cord. Eur J Biochem. 1985;151:625–629. doi: 10.1111/j.1432-1033.1985.tb09150.x. [DOI] [PubMed] [Google Scholar]
- 22.Kahya N, Scherfeld D, Bacia K, Schwille P. Lipid domain formation and dynamics in giant unilamellar vesicles explored by fluorescence correlation spectroscopy. J Struct Biol. 2004;147:77–89. doi: 10.1016/j.jsb.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 23.Kahya N, Schwille P. Fluorescence correlation studies of lipid domains in model membranes. Mol Membr Biol. 2006;23:29–39. doi: 10.1080/09687860500489099. [DOI] [PubMed] [Google Scholar]
- 24.Hess ST, Huang S, Heikal AA, Webb WW. Biological and chemical applications of fluorescence correlation spectroscopy: a review. Biochemistry. 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- 25.Vlad M, Segal E. A kinetic analysis of Langmuir model for adsorption within the framework of Jovanovic theory; A generalization of the Jovanovic isotherm. Surface Sci. 1979;79:608–616. [Google Scholar]
- 26.Andree HA, Reutelingsperger CP, Hauptmann R, Hemker HC, Hermens WT, Willems GM. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J Biol Chem. 1990;265:4923–4928. [PubMed] [Google Scholar]
- 27.Follenius-Wund A, Piémont E, Freyssinet JM, Gérard D, Pigault C. Conformational adaptation of annexin V upon binding to liposomes: a time-resolved fluorescence study. Biochem Biophys Res Commun. 1997;234:111–116. doi: 10.1006/bbrc.1997.6596. [DOI] [PubMed] [Google Scholar]
- 28.Rand JH, Wu XX, Quinn AS, Chen PP, McCrae KR, Bovill EG, Taatjes DJ. Human monoclonal antiphospholipid antibodies disrupt the annexin A5 anticoagulant crystal shield on phospholipid bilayers: evidence from atomic force microscopy and functional assay. Am J Pathol. 2003;163:1193–1200. doi: 10.1016/S0002-9440(10)63479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter RP, Him JL, Tessier B, Tessier C, Brisson AR. On the kinetics of adsorption and two-dimensional self-assembly of annexin A5 on supported lipid bilayers. Biophys J. 2005;89:3372–3385. doi: 10.1529/biophysj.105.064337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andree HA, Stuart MC, Hermens WT, Reutelingsperger CP, Hemker HC, Frederik PM, Willems GM. Clustering of lipid-bound annexin V may explain its anticoagulant effect. J Biol Chem. 1992;267:17907–17912. [PubMed] [Google Scholar]
- 31.Reviakine II, Bergsma-Schutter W, Brisson A. Growth of protein 2-D crystals on supported lipid bilayers imaged in situ by AFM. J Struct Biol. 1998;121:356–361. doi: 10.1006/jsbi.1998.4003. [DOI] [PubMed] [Google Scholar]
- 32.Pigault C, Follenius-Wund A, Schmutz M, Freyssinet JM, Brisson A. Formation of two-dimensional arrays of annexin V on phosphatidylserine-containing liposomes. J Mol Biol. 1994;236:199–208. doi: 10.1006/jmbi.1994.1129. [DOI] [PubMed] [Google Scholar]
- 33.Sabra MC, Uitdehaag JC, Watts A. General model for lipid-mediated two-dimensional array formation of membrane proteins: application to bacteriorhodopsin. Biophys J. 1998;75:1180–1188. doi: 10.1016/S0006-3495(98)74037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hess ST, Sheets ED, Wagenknecht-Wiesner A, Heikal AA. Quantitative analysis of the fluorescence properties of intrinsically fluorescent proteins in living cells. Biophys J. 2003;85:2566–2580. doi: 10.1016/s0006-3495(03)74679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagerholm BC, Weinreb GE, Jacobson K, Thompson NL. Detecting microdomains in intact cell membranes. Annu Rev Phys Chem. 2005;56:309–336. doi: 10.1146/annurev.physchem.56.092503.141211. [DOI] [PubMed] [Google Scholar]
- 36.Han JJ, Boo DW. Reversible immobilization of diffusive membrane-associated proteins using a liquid–gel bilayer phase transition: a case study of annexin V monomers. Langmuir. 2009;25:3083–3088. doi: 10.1021/la803903j. [DOI] [PubMed] [Google Scholar]
- 37.Gilmanshin R, Creutz CE, Tamm LK. Annexin IV reduces the rate of lateral lipid diffusion and changes the fluid phase structure of the lipid bilayer when it binds to negatively charged membranes in the presence of calcium. Biochemistry. 1994;33:8225–8232. doi: 10.1021/bi00193a008. [DOI] [PubMed] [Google Scholar]
- 38.Weiss M, Hashimoto H, Nilsson T. Anomalous protein diffusion in living cells as seen by fluorescence correlation spectroscopy. Biophys J. 2003;84:4043–4052. doi: 10.1016/S0006-3495(03)75130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saxton MJ. Anomalous diffusion due to obstacles: a Monte Carlo study. Biophys J. 1994;66:394–401. doi: 10.1016/s0006-3495(94)80789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loura LMS, Ramalho JPP. Location and dynamics of acyl chain NBD-labeled phosphatidylcholine (NBD-PC) in DPPC bilayers. A molecular dynamics and time-resolved fluorescence anisotropy study. Biochim Biophys Acta. 2007;1768:467–478. doi: 10.1016/j.bbamem.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 41.Loura LMS, Fernandes F, Fernandes AC, Ramalho JPP. Effects of fluorescent probe NBD-PC on the structure, dynamics and phase transition of DPPC. A molecular dynamics and differential scanning calorimetry study. Biochim Biophys Acta. 2008;1778:491–501. doi: 10.1016/j.bbamem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 42.Huster D, Müller P, Arnold K, Herrmann A. Dynamics of membrane penetration of the fluorescent 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) group attached to an acyl chain of phosphatidylcholine. Biophys J. 2001;80:822–831. doi: 10.1016/S0006-3495(01)76061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huster D, Müller P, Arnold K, Herrmann A. Dynamics of lipid chain attached fluorophore 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) in negatively charged membranes determined by NMR spectroscopy. Eur Biophys J. 2003;32:47–54. doi: 10.1007/s00249-002-0264-9. [DOI] [PubMed] [Google Scholar]
- 44.Elegbede AI, Srivastava DK, Hinderliter A. Purification of recombinant annexins without the use of phospholipids. Protein Expr Purif. 2006;50:157–162. doi: 10.1016/j.pep.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Kyoung M, Karunwi K, Sheets ED. A versatile multi-mode microscope to probe and manipulate nanoparticles and biomolecules. J Microsc. 2007;225:137–146. doi: 10.1111/j.1365-2818.2007.01725.x. [DOI] [PubMed] [Google Scholar]
- 46.Vats K, Kyoung M, Sheets ED. Characterizing the chemical complexity of patterned biomimetic membranes. Biochim Biophys Acta. 2008;1778:2461–2468. doi: 10.1016/j.bbamem.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogt BD, Lin EK, Wu WL, White CC. Effect of film thickness on the validity of the Sauerbrey equation for hydrated polyelectrolyte films. J Phys Chem B. 2004;108:12685–12690. [Google Scholar]
- 48.Ozeki T, Furusawa H, Okahata Y. Evaluation of RNA structures by using a probe-immobilized 27 MHz quartz crystal microbalance. Chem Lett. 2006;35:46–47. [Google Scholar]
- 49.Weast RC. CRC Handbook of Chemistry and Physics. 68. CRC Press; Boca Raton, FL: 1972. p. B-133. [Google Scholar]
- 50.Okahata Y, Niikura K, Furusawa H, Matsuno H. A highly sensitive 27 MHz quartz-crystal microbalance as a device for kinetic measurements of molecular recognition on DNA strands. Anal Sci. 2000;16:1113–1119. [Google Scholar]
- 51.Kastl K, Ross M, Gerke V, Steinem C. Kinetics and thermodynamics of annexin A1 binding to solid-supported membranes: a QCM study. Biochemistry. 2002;41:10087–10094. doi: 10.1021/bi025951z. [DOI] [PubMed] [Google Scholar]
- 52.Elson EL, Magde D. Fluorescence correlation spectroscopy. I Conceptual basis and theory. Biopolymers. 1974;13:1–27. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 53.Heikal AA, Hess ST, Baird GS, Tsien RY, Webb WW. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine) Proc Natl Acad Sci USA. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hac AE, Seeger HM, Fidorra M, Heimburg T. Diffusion in two-component lipid membranes—a fluorescence correlation spectroscopy and Monte Carlo study. Biophys J. 2005;88:317–333. doi: 10.1529/biophysj.104.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bacia K, Scherfeld D, Kahya N, Schwille P. Fluorescence correlation spectroscopy relates rafts in model and native membranes. Biophys J. 2004;87:1034–1043. doi: 10.1529/biophysj.104.040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wawrezinieck L, Rigneault H, Marguet D, Lenne PF. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheets ED, Lee GM, Simson R, Jacobson K. Transient confinement of a glycosylphosphatidylinositol-anchored protein in the plasma membrane. Biochemistry. 1997;36:12449–12458. doi: 10.1021/bi9710939. [DOI] [PubMed] [Google Scholar]