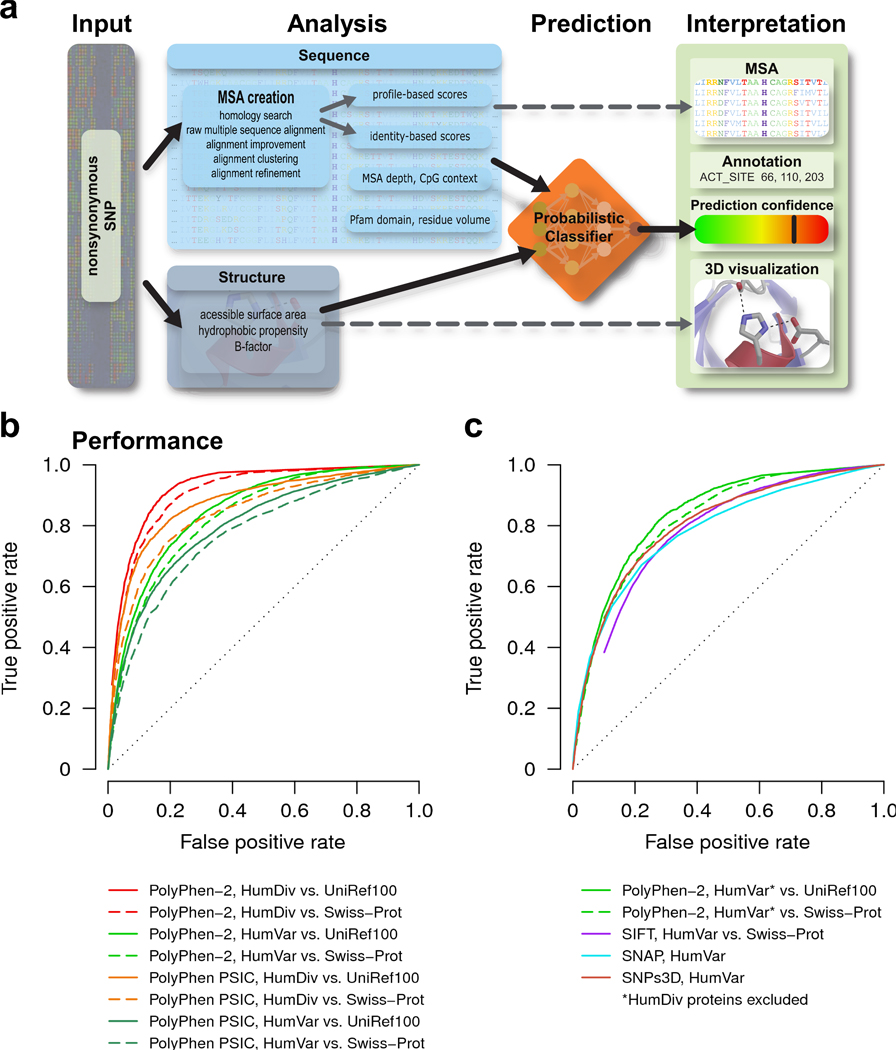
Figure 1.
PolyPhen-2 pipeline and prediction accuracy. (a) Overview of the algorithm. (b) Receiver operating characteristic (ROC) curves for predictions made by PolyPhen-2 using five-fold cross-validation on HumDiv (red) and HumVar3 (light green). UniRef100 (solid lines) and Swiss-Prot (dashed lines) databases were used for the homology search in the sequence analysis pipeline. Also shown are corresponding ROC curves for PolyPhen on HumDiv (orange) and HumVar (dark green) calculated from the difference between PSIC scores1 of the wild type and the mutant amino acid residues. (c) ROC curves for PolyPhen-2 trained on HumDiv and tested on a subset of HumVar non-overlapping with HumDiv (green). UniRef100 (solid lines) and Swiss-Prot (dashed lines) databases were used for the homology search. Also shown are ROC curves for SIFT4 (blue), SNAP5 (cyan) and SNPs3D6 (brown) on HumVar. Methods other than PolyPhen-2 and PolyPhen could not easily be applied to HumDiv because using the same sequences for obtaining both multiple alignments and non-damaging replacements must be avoided. SIFT was used in conjunction with Swiss-Prot database, SNAP and SNPs3D were used with their corresponding default databases. We used SIFT with Swiss-Prot database for homology search since Swiss-Prot does not contain incomplete sequences, sequences of splice forms and sequences of human allelic variants, making it possible to guarantee that allelic variants used in testing datasets would not appear in multiple sequence alignments used in computing prediction rules by other methods.