Figure 4.
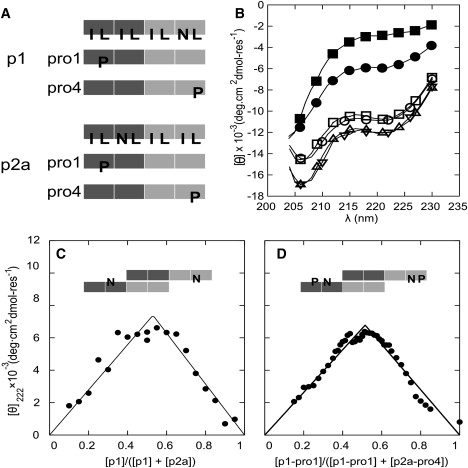
Proline-scanning mutagenesis. (A) Schematics of the proline-mutant peptides. The light and dark blocks represent oppositely charged heptad repeats. In the uppermost schematics, I, L, and N indicate isoleucine, leucine, and asparagine residues at the H-type sites in the HPPHPPP repeats of the parent sequences. In the remaining schematics, P highlights proline residues that have replaced specific L residues, and all of the other H sites remain unchanged. (B) CD spectra from mixtures of proline variants and parent peptides: p1-Pro1:p2a (■), p1:p2a-Pro4 (•), p1:p2a (○), p1-Pro4:p2a (▵), p1:p2a-Pro1, (▿), and p1-Pro4:p2a-Pro1 (□). (C) Job plot for mixtures of SAF-p1 and SAF-p2a, showing a 1:1 binding stoichiometry. (D) Similar Job plot for mixtures of SAF-p1-pro4 and SAF-p2a-pro1, showing an unaltered binding stoichiometry. In both C and D, the total peptide concentration was kept at 200 μM, and lines are fits to the data as described in the Supporting Material.
