Abstract
Store-operated calcium entry (SOCE) is a key evolutionarily conserved process whereby decreases in endoplasmic reticulum Ca2+ content lead to the influx of Ca2+ across the plasma membrane. How this process is regulated in specific tumor cell types is poorly understood. In an effort to address this concern, we obtained and tested primary Wilms tumor cells, finding no detectable SOCE in this cell type. Analysis of the expression levels of STIM1 and ORAI1 (the molecular mediators of SOC) revealed poor STIM1 expression. Analysis of the STIM1 promoter using the TESS search system (University of Pennsylvania) revealed four putative response elements to the zinc-finger proteins WT1 (Wilms tumor suppressor 1) and EGR1 (early growth response 1). Either overexpression of WT1 or knockdown of EGR1 resulted in loss of STIM1 expression and a resultant decrease in SOCE. Furthermore, examination of Egr1 knock-out animals revealed loss of STIM1 expression in multiple tissues. Finally, using chromatin immunoprecipitation, we reveal direct binding of both WT1 and EGR1 to putative response elements located within 500 bp of the transcriptional start site of STIM1. Considering that WT1 and EGR1 are well described oncogenes and tumor suppressors, these observations may reveal new mechanisms responsible for distinct Ca2+ signals in cancer cells.
Keywords: Oncogene, Tumor/Suppressor, Calcium Channels, Transcription Promoter, Transcription Regulation, EGR1, STIM1, WT1
Introduction
Changes in cytosolic Ca2+ levels are a common component of the signal transduction pathways for numerous growth factors and cytokines. Thus, activation of phospholipase C-coupled receptors (primarily via either G protein or tyrosine kinase receptors) results in the generation of the second messenger inositol 1,4,5-trisphosphate (1). Inositol 1,4,5-trisphosphate diffuses rapidly through the cytosol and interacts with its receptor on the endoplasmic reticulum (ER),2 resulting in both transient ER Ca2+ release and a lengthy increased influx of Ca2+ across the plasma membrane, a process termed capacitative or store-operated Ca2+ entry (SOCE) (2). SOCE has been shown to regulate numerous fundamental processes in cell biology, including migration (3, 4), proliferation (4–6), and differentiation (7, 8). Given the impact of these pathways on cancer cell biology, it is not surprising that altered Ca2+ signaling can be observed in numerous classes of cancer cells. Indeed, inhibition of either the phosphatidylinositol pathway (9) or calcium influx (10, 11) can induce either growth arrest or cell death in a variety of tumor cells. Despite these observations, Ca2+ signals remain poorly utilized therapeutic targets. This is in part because these studies were all performed prior to the discovery of the identities of the molecular mediators of SOCE. Without this insight, it was not possible to link changes in Ca2+ signals with changes in the expression and function of oncogenes and tumor suppressors that cause tumor formation.
After nearly 20 years of investigations into the mysteries of SOCE, the identities of the key molecular components of this process have finally been revealed (for recent reviews, see Refs. 12 and 13). Thus, the type 1A transmembrane protein STIM1 serves a dual role as an ER Ca2+ sensor and activator of SOCE (14, 15), whereas the plasma membrane-localized transmembrane protein ORAI1 is the store-operated Ca2+ channel (16–18). Both of these proteins have mammalian homologues (STIM2, ORAI2, and ORAI3) that do not appear to be required for SOCE (4, 15, 19, 20) and likely serve modulatory roles in related processes. Despite an exciting recent report that SOCE is a critical and required component of breast tumor cell migration and metastasis (3), there remains no published insight into the regulation of SOCE components at the level of transcription or their relationship to tumorigenesis.
In early investigations performed prior to the discovery of its role in Ca2+ signaling, STIM1 was described as a tumor suppressor because it causes growth arrest in human G401 rhabdoid tumor cells (21, 22), a kidney-derived rhabdoid cell line often used to study chromosomal changes in Wilms tumor (23) due to the fact that they lack expression of WT1 (Wilms tumor suppressor 1). WT1 is a zinc-finger transcription factor that regulates the expression of multiple growth factors such as colony-stimulating factor (24), insulin-like growth factor I (25), and platelet-derived growth factor (26). Thus, loss of WT1 leads to up-regulation of these growth factors and the formation of Wilms tumor, leading to its original classification as a tumor suppressor (27). However, subsequent studies have revealed a potential role for WT1 as an oncogene because it is up-regulated in a variety of human cancers such as astrocytic tumors (28), breast cancer (29), leukemia (30), and sporadic Wilms tumor (31, 32), which accounts for ∼85% of all Wilms tumors. WT1 is a member of the EGR (early growth response) family, primarily due to similarities in the consensus sequences of WT1 and EGR1. However, the consequences of binding are most often mutually opposing; EGR1 activates the transcription of genes that WT1 represses (33, 34). Like WT1, EGR1 expression levels are atypical in multiple neoplastic cell types such as prostate cancer (35), glioblastoma (36), and Wilms tumor (32). Furthermore, EGR1 can also act as either an oncogene (35) or a tumor suppressor (37) in different cell types. In this work, we reveal that STIM1 expression is directly regulated by EGR1 and WT1, with EGR1 driving STIM1 expression and WT1 antagonizing this effect. Given the well described impact of these oncogenes/tumor suppressors on tumorigenesis, these findings provide important new insight into differential Ca2+ signaling in cancer cells.
EXPERIMENTAL PROCEDURES
Cell Culture
G401 cells were maintained in McCoy's medium (10% fetal bovine serum). Human embryonic kidney 293 (HEK293) cells were maintained in Dulbecco's modified Eagle's medium (10% fetal bovine serum); for HEK293 cells stably expressing STIM1 (20), Dulbecco's modified Eagle's medium was also supplemented with G418. Human kidney 2 (HK-2) cells were maintained in serum-free keratinocyte medium (Invitrogen). Rat pheochromocytoma 12 (PC12) cells were maintained in Dulbecco's modified Eagle's medium (10% horse serum, 5% fetal bovine serum). All media were supplemented with antibiotics and maintained at 37 °C with 5% CO2.
Wilms Tumor Isolation
Isolated primary Wilms tumors were washed in phosphate-buffered saline and cut into small pieces. These were then incubated with collagenase V (1 mg/ml) at 37 °C with agitation for 15 min. The mixture was filtered through a mesh screen into Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and centrifuged at 300 × g for 5 min. The pellet was resuspended in phosphate-buffered saline supplemented with bovine serum albumin and 2 mm EDTA. Rat anti-mouse major histocompatibility complex II antibody (Serotec) was added to this solution (1 μg of antibody/50,000 cells) and incubated at 4 °C for 20 min. Cells were then centrifuged at 300 × g for 5 min, washed with phosphate-buffered saline, and resuspended in phosphate-buffered saline supplemented with 0.1% bovine serum albumin and 2 mm EDTA. Dynabeads (Invitrogen) were added for 30 min at 4 °C with agitation and precipitated with a DynaMag-2 magnet (Invitrogen). Human primary Wilms tumor cells were then collected from the supernatant and either plated onto coverslips for Ca2+ measurement or lysed for Western blot analysis.
WT1 and EGR1 Constructs and Transfections
WT1 was obtained from Dr. Dan Liebermann (Temple University). Human EGR1 was generously provided by Dr. Kenneth Tew (Medical University of South Carolina). Validated small interfering RNA (siRNA) sequences targeting human WT1 (position 1423, GGACUGUGAACGAAGGUUUtt; position 2598, GGAUCUCCACUGAUAAGACtt) along with a control sequence (GGUUCUCCACCUUUAUAGGUGGCUU) were obtained from Applied Biosystems. Stealth EGR1 siRNAs (position 854, CAUCCAACGACAGCAGUCCCAUUUA; position 1851, GCUUUCGGACAUGACAGCAACCUUU) were designed using the Block-it siRNA designer (Invitrogen). DNA constructs and RNA sequences were introduced by electroporation using the Gene Pulser Xcell electroporation system (Bio-Rad) at 220 V and 500 microfarads (25-ms pulse length), followed by 48 h in culture.
Cytosolic Ca2+ Measurements
Ratiometric imaging of intracellular Ca2+ using Fura-2/acetoxymethyl ester was performed as described previously (38). Cells grown on coverslips were placed in cation-safe solution (107 mm NaCl, 7.2 mm KCl, 1.2 mm MgCl2, 11.5 mm glucose, 20 mm Hepes-NaOH, pH 7.2) and loaded with Fura-2/acetoxymethyl ester (2 μm) for 30 min at 24 °C. Cells were washed, and dye was allowed to de-esterify for a minimum of 30 min at 24 °C. Approximately 85% of the dye was confined to the cytoplasm as determined by the signal remaining after saponin permeabilization (39). Ca2+ measurements were made using a Leica DMI 6000B fluorescence microscope controlled by Slidebook software (Intelligent Imaging Innovations, Denver, CO). Fluorescence emission at 505 nm was monitored while alternating between 340- and 380-nm excitation wavelengths at a frequency of 0.67 Hz; intracellular Ca2+ measurements are shown as 340/380 nm ratios obtained from groups (35–45) of single cells.
RNA Extraction and Reverse Transcription
RNA was collected via phenol/chloroform extraction. Briefly, cells were lysed with TRIzol reagent (Molecular Research Center, Inc.). Chloroform was then added to induce phase separation. After extraction of RNA from the aqueous phase by ethanol precipitation, first-strand cDNA synthesis was completed by incubating RNA at 55 °C (30 min) with random primers and reverse transcriptase (Applied Biosystems).
Quantitative PCR
Real-time PCR was performed on an Applied Biosystems 7300 real-time PCR system. 600 ng of cDNA was added with primers (500 nm final concentration) and SYBR Green MasterMix (Applied Biosystems). PCR was performed for 5 min at 95 °C, followed by 40 cycles between 95 °C (15 s) and 55–65 °C (varying depending on the primer; 1 min). The cycle threshold for the cDNA of interest (a) was normalized to the cycle threshold of TATA-binding protein (b) in the following formula: 2̂(a − b).
Sample Collection and Western Blot Analysis
Cells were lysed in Nonidet P-40 lysis buffer (1% (w/v) Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 8.0, with protease inhibitors), cleared by centrifugation, and normalized for protein content. When protein were extracted from mice, tissues were homogenized and resuspended in Nonidet P-40 lysis buffer. Homogenate was freeze-thawed three times prior to clearing by centrifugation and normalization for protein content. Proteins were resolved on 6–8% SDS-polyacrylamide gels, transferred to nitrocellulose paper, and analyzed with the indicated antibodies as described previously (40).
Genotyping
Mouse tails were clipped (0.5 cm) and incubated overnight in lysis buffer (100 mm Tris, pH 8.5, 5 mm EDTA, 200 mm NaCl, 0.2% SDS, 10 mg/ml proteinase K) at 50 °C, cleared by centrifugation, and genotyped by PCR. PCR was performed for 5 min at 95 °C, followed by 30 cycles at 95 °C for 1 min, 60 °C for 14 s, and 72 °C for 1.5 min.
Chromatin Immunoprecipitation (ChIP)
ChIP kits were obtained from Millipore and used according to the manufacturer's instructions. Briefly, cells were treated with 1% formaldehyde for 10 min, followed by lysis in SDS buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.1). Lysate was then sheared by sonication (4 × 10-s pulses at 30% power), followed by immunoprecipitation with anti-WT1, anti-EGR1, or rabbit IgG control antibodies. Immunoprecipitated DNA was then heated (65 °C) in 25 mm NaCl (4 h) to eliminate cross-links and analyzed by PCR. PCR was performed for 10 min at 95 °C, followed by 40 cycles at 95 °C (15 s), 60 °C (45 s), and 70 °C (15 s) (varying depending on the primer; 1 min). PCR products were then separated by PAGE (8%) and visualized using ethidium bromide staining at the end of each experiment.
Materials
Peptide affinity-purified specific antibodies to STIM1 were produced by 21st Century Biochemicals (Marlboro, MA). ChIP-grade anti-WT1 and anti-EGR1 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit IgG was from Bethyl Laboratories (Montgomery, TX). Anti-ORAI1 antibody was from Sigma. Fura-2/acetoxymethyl ester and horseradish peroxidase-conjugated goat anti-rabbit and rabbit anti-mouse antibodies were from Invitrogen. All restriction enzymes were from New England Biosciences. Thapsigargin was from EMD Biosciences.
RESULTS AND DISCUSSION
SOCE in Wilms Tumor
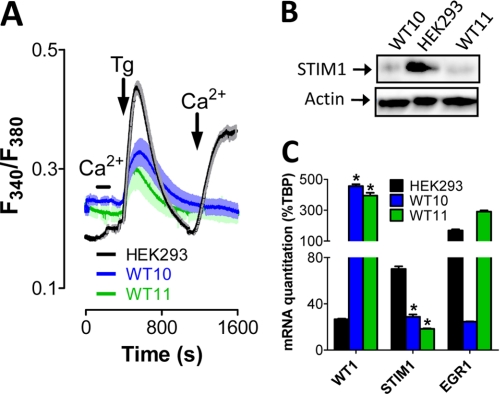
To determine whether changes in SOCE are a common component of Wilms tumor, we examined cells extracted from two Wilms tumor explants termed WT10 and WT11. These are human Wilms tumors propagated subcutaneously in SCID (severe combined immunodeficiency) mice that have been successfully used as a model for sporadic Wilms tumor (41, 42). These tumors were removed from the mice, dissociated, separated from host cells, and loaded with Fura-2 to measure changes in cytosolic Ca2+ concentration. Interestingly, depleting ER Ca2+ content via the addition of thapsigargin failed to stimulate SOCE in either Wilms tumor sample (Fig. 1A). By comparison, a similar manipulation in HEK293 cells (a well described kidney-derived cell line) led to easily detectable SOCE (Fig. 1A). In an effort to identify potential causes of loss of SOCE in Wilms tumor cells, we examined the expression levels of STIM1 and ORAI1, the primary molecular mediators of SOCE. Western blot analysis of protein lysates from both WT10 and WT11 cells revealed significant loss of STIM1 protein in comparison with HEK293 cells (Fig. 1B), whereas ORAI1 expression levels determined by quantitative PCR were highly variable (supplemental Fig. S1). Hence, there was a clear correlation between loss of SOCE and STIM1 but not ORAI1 expression in Wilms tumor cells.
FIGURE 1.
Loss of SOC in Wilms tumor cells. A, SOCE was measured in Fura-2-loaded WT10, WT11, and HEK293 cells after ER Ca2+ depletion via the addition of the SERCA inhibitor thapsigargin (Tg; 2 μm) in nominally Ca2+-free medium. Extracellular Ca2+ concentration was increased from 0 to 1 mm either before or after store depletion where indicated to differentiate between store-independent (before thapsigargin) and store-dependent (after thapsigargin) Ca2+ entry. Shaded areas indicate S.E. B, protein extracted from WT10, WT11, and HEK293 cells was analyzed by Western blotting for STIM1 expression. actin was used as a loading control. C, RNA extracted from WT10, WT11, and HEK293 cells was analyzed by quantitative PCR for expression of TATA-binding protein (TBP), STIM1, WT1, and EGR1. mRNA expression levels are shown as %TBP. *, significant differences from expression levels in HEK293 cells as determined by analysis of variance with Tukey's post-hoc test (p < 0.05). All experiments were completed a minimum of three times.
As discussed in the Introduction, expression levels of both WT1 and EGR1 are often aberrant in Wilms tumors (31, 32). Therefore, we examined their expression levels to assess any potential correlations with STIM1 expression levels. Interestingly, WT1 expression was increased >10-fold at the mRNA level over HEK293 cells in both Wilms tumors, whereas STIM1 expression was decreased ∼2-fold (Fig. 1C). In contrast, EGR1 expression was highly variable. This led us to speculate that WT1 may negatively regulate STIM1 expression. In support of this concept, analysis of the genomic DNA sequence 11p15.5 in the region immediately upstream of the STIM1 the TESS search system (University of Pennsylvania) revealed four putative response elements for both WT1 and EGR1 within 500 bp of the STIM1 transcriptional start site. Therefore, we examined the impact of these transcription factors on STIM1 expression and function.
Control of STIM1 Expression and Activation by WT1 and EGR1
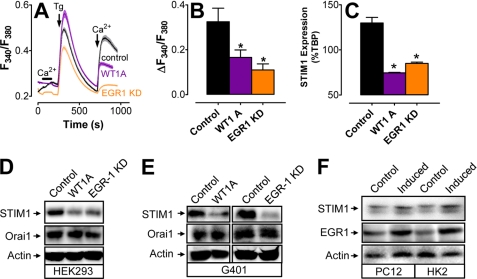
Given the observed correlations between WT1 and both SOCE and STIM1 expression in Wilms tumor cells and the existence of putative WT1/EGR1-binding sites in the STIM1 promoter, we predicted that either overexpression of WT1 or knockdown of EGR1 would lead to decreases in both SOCE and STIM1 expression. Due to our inability to culture Wilms tumor cells, we were unable to perform this experiment in the WT10 or WT11 model system. However, HEK293 cells can be transfected at high efficiency with either cytomegalovirus-driven expression plasmids or targeted siRNA sequences (supplemental Fig. S2). Furthermore, significant decreases in SOCE (Fig. 2, A and B), STIM1 mRNA (Fig. 2C), and STIM1 protein (Fig. 2D) were observed in HEK293 cells transfected with either WT1A (a major DNA-binding WT1 splice variant) or EGR1 siRNA. TATA-binding protein was used as an internal control for quantitative PCR, whereas ORAI1 and actin were used as loading controls for Western blot analysis. An analogous experiment in G401 cells revealed a similar loss of STIM1 expression after transfection with either WT1A or EGR1 siRNA with no changes in either ORAI1 or actin expression levels (Fig. 2E). Finally, in an effort to determine whether the WT1/EGR1-dependent changes in SOCE were due solely to loss of STIM1, this experiment was repeated in HEK293 cells stably expressing STIM1 (supplemental Fig. S3) (20). In this case, neither WT1A overexpression nor EGR1 knockdown had any effect on SOCE. Hence, loss of STIM1 expression is sufficient to account for changes in SOCE associated with the level of expression of WT1 or EGR1. The fact that STIM1 and WT1 had an inverse relationship is consistent with the inverse correlation we observed in Wilms tumor cells (Fig. 1); in contrast, we did not observe any relationship between EGR1 and STIM1 expression in Wilms tumor cells. We were somewhat surprised by these differences; however, prior studies have shown that, despite highly similar binding characteristics, differences in WT1/EGR1 response elements can greatly affect their relative affinities (43). If so, our observation would be consistent with a dominant role in suppression of STIM1 expression by WT1.
FIGURE 2.
Control of STIM1 expression by WT1 and EGR1. A, representative traces of Ca2+ responses in Fura-2-loaded HEK293 cells after overexpression of WT1A or knockdown of EGR1. Shaded areas indicate S.E. B, quantitation of the change in cytosolic Ca2+ concentration after store depletion from three experiments performed as depicted in A. C, STIM1 expression levels in HEK293 cells transfected with either WT1A- or EGR1-targeted siRNA as determined by quantitative PCR. D, Western blot analysis of STIM1, ORAI1, and actin expression in HEK293 cells transfected with WT1A or EGR1 siRNA. KD, knockdown. E, Western blot analysis of STIM1, ORAI1, and actin expression in G401 cells transfected with WT1A or EGR1 siRNA. F, Western blot analysis of STIM1, EGR1, and actin expression in pheochromocytoma 12 or human kidney 2 cells after incubation in either full or 10% growth medium for 48 h. *, significant differences from the control (p < 0.05) as determined by one-way analysis of variance with Tukey's post-hoc test.
If WT1 inhibits STIM1 expression and EGR1 drives STIM1 expression, then we predicted that either knocking down WT1 or overexpressing EGR1 in HEK293 cells would increase STIM1 expression. However, that prediction proved to be incorrect (supplemental Fig. S4). Although we cannot definitively explain this observation, the likely reason for this is that endogenous STIM1 expression is at maximal levels in HEK293 cells under unmanipulated conditions. Indeed, because EGR1 expression can be induced with serum, we compared both EGR1 and STIM1 expression in cells that had either been serum-deprived or maximally stimulated. Although serum deprivation had no effect on either STIM1 or EGR1 expression in HEK293 cells (data not shown), this manipulation significantly increased both STIM1 and EGR1 expression in both PC12 and HK-2 cells (Fig. 2F). Hence, induction of EGR1 is sufficient to increase STIM1 expression if cells are not maintained under optimal growth conditions.
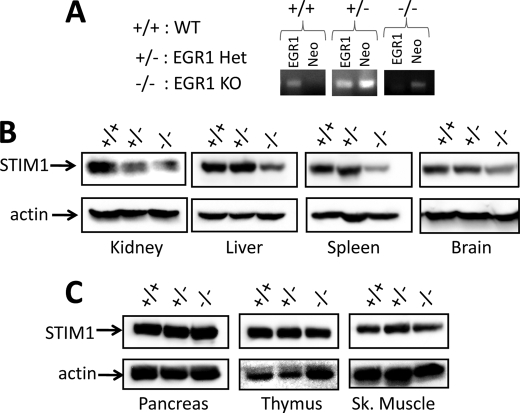
Analysis of Egr1 Knock-out Tissues
Although loss of Wt1 is embryonic lethal (44), Egr1 knock-out mice were generated in the Milbrandt laboratory over 15 years ago (45) and are readily available. Therefore, in an effort to determine whether or not EGR1-mediated control of STIM1 expression is required in vivo, we examined tissues obtained from these Egr1+/+, Egr1+/−, and Egr1−/− animals (Fig. 3A). Intriguingly, kidney (source of Wilms tumor) homogenates from either Egr1 heterozygous or knock-out animals exhibited a significant decrease in STIM1 expression (Fig. 3B). In contrast, STIM1 expression was normal in all other Egr1 heterozygous tissues examined. However, significant decreases in STIM1 expression were observed in liver, spleen, and brain homogenates (Fig. 3B) but not pancreas, thymus, or skeletal muscle homogenates (Fig. 3C) obtained from Egr1 knock-out mice. These observations indicate that EGR1-dependent expression of STIM1 is highly tissue-specific, with kidney exhibiting the greatest dependence on EGR1 for STIM1 expression, followed by liver, spleen, and brain. Nevertheless, the fact that loss of STIM1 expression was neither universal nor complete indicates that STIM1 expression is likely dependent on multiple transcription factors, the identities of which have not yet been determined.
FIGURE 3.
Analysis of STIM1 expression in Egr1 knock-out mouse tissues. A, DNA was extracted from C57Bl/6 mouse tails, amplified by PCR, and analyzed by PAGE. B and C, protein was extracted from Egr1+/+, Egr1+/−, and Egr1−/− tissues exhibiting EGR1-dependent STIM1 expression (B; kidney, spleen, liver, and brain) and those exhibiting EGR1-independent STIM1 expression (C; pancreas, thymus, and skeletal muscle). STIM1 expression was determined by Western blot analysis, whereas actin expression was determined as a loading control. WT, wild type; Het, heterozygous; KO, knock-out; Sk., skeletal.
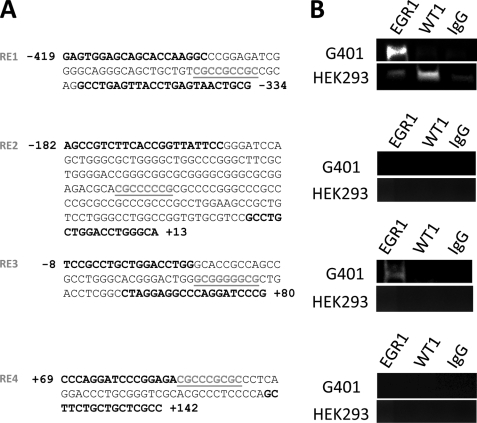
WT1 and EGR1 Response Elements within the STIM1 Promoter
On the basis of the studies described above, we were highly confident that STIM1 expression is under the control of WT1 and EGR1. Therefore, we analyzed regions of the human and murine genomes located in the vicinity of the STIM1 transcriptional start site using the TESS search system (University of Pennsylvania). Intriguingly, four highly conserved WT1/EGR1 response elements (REs) were located within 500 bp of the start of the STIM1 gene in both the human and mouse sequences (76.3% homology for ∼700-bp regions analyzed). In an effort to distinguish between direct and indirect modes of regulation, we performed ChIP to determine which (if any) of these putative REs exhibit bona fide WT1/EGR1 binding. This was achieved using either ChIP-grade rabbit anti-EGR1 and anti-WT1 antibodies or control rabbit IgG to pull down DNA extracted from both G401 cells (WT1-null) (23) and HEK293 cells. Identification of successful binding was achieved by PCR using primers flanking each of the four putative REs (Fig. 4A), followed by PAGE. Interestingly, the strongest amplifications of putative RE1 were observed using either anti-EGR1 antibodies in G401 extracts or anti-WT1 antibodies in HEK293 extracts but not when using either rabbit IgG or unbound beads (Fig. 4B, top). In contrast, pulldown of RE1 using anti-EGR1 antibodies in HEK293 extracts was comparable with rabbit IgG pulldown, presumably reflecting enhanced affinity for WT1 over EGR1 for binding to this response element. Although both RE2 and RE4 were negative for interaction with either EGR1 or WT1, we did observe pulldown of RE3 using anti-EGR1 antibodies in G401 extracts only. The reasons for the cell-type specificity of EGR1 interactions with RE3 are not immediately clear but may reflect the presence of other as yet unidentified transcription factors involved in the control of STIM1 expression. Nevertheless, these observations provide strong support for the concept that EGR1 and WT1 regulate STIM1 expression via direct interactions with RE1.
FIGURE 4.
WT1 and EGR1 bind to the genomic region adjacent to the STIM1 transcriptional start site. A, analysis of the DNA sequence of 11p15.5 in the region surrounding the STIM1 transcriptional start site revealed four putative WT1/EGR1 REs (underlined). B, binding of WT1 and EGR1 to the four putative response elements was determined by ChIP of genomic DNA extracted from either HEK293 or G401 cells. Protein-DNA complexes were immunoprecipitated with anti-EGR1 or anti-WT1 antibodies or anti-rabbit IgG. DNA was amplified by PCR using the primers depicted in boldface in A. No template was used as a negative control for each PCR. Each experiment was completed a minimum of three times.
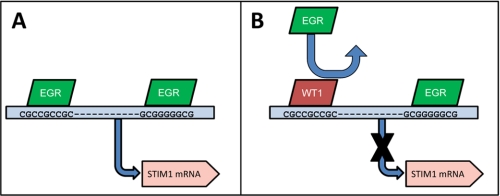
Conclusions
In this work, we provide evidence that two prominent oncogenes/tumor suppressors directly regulate STIM1 expression. Based on our findings, we propose that EGR1 binds to RE1 and potentially RE3 (depicted in Fig. 5). Furthermore, we suggest that WT1 blocks EGR1-dependent STIM1 expression by interfering with EGR1 binding to RE1. To our knowledge, this is the first study proposing the involvement of any specific transcription factors in the control of STIM1 expression. However, the fact that WT1 and EGR1 are so intimately involved in tumorigenesis has led us to speculate that WT1/EGR1-dependent control of STIM1 expression may be responsible for some of the differences in Ca2+ signaling that have been reported in numerous cancer types. Although the nature of the contribution of aberrant STIM1 expression to carcinogenesis and/or tumor progression remains unclear, this important question will be addressed in future studies. Nevertheless, given numerous studies linking dysregulation of Ca2+ homeostasis and apoptosis, WT1 and EGR1 may have untapped potential as biomarkers for the design of future therapeutic strategies targeting SOCE in cancer cells.
FIGURE 5.
Model for control of STIM1 expression by WT1 and EGR1. Putative EGR1 response elements are depicted relative to the start site for transcription of STIM1 mRNA (marked by the blue arrow). Inhibition of EGR1 binding at RE1 is marked with an arrow.
Supplementary Material
Acknowledgments
We thank Drs. Dan Liebermann and Barbara Hoffman (Temple University) for providing the EGR1 knock-out mice. In addition, we thank Dr. Ana Gamero (Temple University) for use of the fluorometer. Finally, we thank Drs. Dale Haines and Muniswamy Madesh (Temple University) for critical reading of the manuscript.
This work was supported by a grant from the Pennsylvania Department of Health (to J. S.) and by Grant 0730184N from the American Heart Association (to J. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- ER
- endoplasmic reticulum
- RE
- response element
- SOCE
- store-operated Ca2+ entry
- ChIP
- chromatin immunoprecipitation
- siRNA
- small interfering RNA.
REFERENCES
- 1.Berridge M. J., Bootman M. D., Roderick H. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 517–529 [DOI] [PubMed] [Google Scholar]
- 2.Venkatachalam K., van Rossum D. B., Patterson R. L., Ma H. T., Gill D. L. (2002) Nat. Cell Biol. 4, E263–E272 [DOI] [PubMed] [Google Scholar]
- 3.Yang S., Zhang J. J., Huang X. Y. (2009) Cancer Cell 15, 124–134 [DOI] [PubMed] [Google Scholar]
- 4.Potier M., Gonzalez J. C., Motiani R. K., Abdullaev I. F., Bisaillon J. M., Singer H. A., Trebak M. (2009) FASEB J. 23, 2425–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdullaev I. F., Bisaillon J. M., Potier M., Gonzalez J. C., Motiani R. K., Trebak M. (2008) Circ. Res. 103, 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Boustany C., Bidaux G., Enfissi A., Delcourt P., Prevarskaya N., Capiod T. (2008) Hepatology 47, 2068–2077 [DOI] [PubMed] [Google Scholar]
- 7.Darbellay B., Arnaudeau S., König S., Jousset H., Bader C., Demaurex N., Bernheim L. (2009) J. Biol. Chem. 284, 5370–5380 [DOI] [PubMed] [Google Scholar]
- 8.Gwack Y., Srikanth S., Oh-Hora M., Hogan P. G., Lamperti E. D., Yamashita M., Gelinas C., Neems D. S., Sasaki Y., Feske S., Prakriya M., Rajewsky K., Rao A. (2008) Mol. Cell. Biol. 28, 5209–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powis G., Gallegos A., Abraham R. T., Ashendel C. L., Zalkow L. H., Grindey G. B., Bonjouklian R. (1994) Cancer Chemother. Pharmacol. 34, 344–350 [DOI] [PubMed] [Google Scholar]
- 10.Cole K., Kohn E. (1994) Cancer Metastasis Rev. 13, 31–44 [DOI] [PubMed] [Google Scholar]
- 11.Soboloff J., Zhang Y., Minden M., Berger S. A. (2002) Exp. Hematol. 30, 1219–1226 [DOI] [PubMed] [Google Scholar]
- 12.Deng X., Wang Y., Zhou Y., Soboloff J., Gill D. L. (2009) J. Biol. Chem. 284, 22501–22505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luik R. M., Lewis R. S. (2007) Trends Mol. Med. 13, 103–107 [DOI] [PubMed] [Google Scholar]
- 14.Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S. H., Tanasa B., Hogan P. G., Lewis R. S., Daly M., Rao A. (2006) Nature 441, 179–185 [DOI] [PubMed] [Google Scholar]
- 17.Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. (2006) Science 312, 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S. L., Yeromin A. V., Zhang X. H., Yu Y., Safrina O., Penna A., Roos J., Stauderman K. A., Cahalan M. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9357–9362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bird G. S., Hwang S. Y., Smyth J. T., Fukushima M., Boyles R. R., Putney J. W. (2009) Curr. Biol. 19, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soboloff J., Spassova M. A., Hewavitharana T., He L. P., Xu W., Johnstone L. S., Dziadek M. A., Gill D. L. (2006) Curr. Biol. 16, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 21.Sabbioni S., Barbanti-Brodano G., Croce C. M., Negrini M. (1997) Cancer Res. 57, 4493–4497 [PubMed] [Google Scholar]
- 22.Sabbioni S., Veronese A., Trubia M., Taramelli R., Barbanti-Brodano G., Croce C. M., Negrini M. (1999) Cytogenet. Cell Genet. 86, 214–218 [DOI] [PubMed] [Google Scholar]
- 23.Garvin A. J., Re G. G., Tarnowski B. I., Hazen-Martin D. J., Sens D. A. (1993) Am. J. Pathol. 142, 375–380 [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington M. A., Konicek B., Song A., Xia X. L., Fredericks W. J., Rauscher F. J., 3rd (1993) J. Biol. Chem. 268, 21271–21275 [PubMed] [Google Scholar]
- 25.Sarfstein R., Werner H. (2006) J. Neurochem. 99, 818–826 [DOI] [PubMed] [Google Scholar]
- 26.Wang Z. Y., Madden S. L., Deuel T. F., Rauscher F. J., 3rd (1992) J. Biol. Chem. 267, 21999–22002 [PubMed] [Google Scholar]
- 27.Little M., Wells C. (1997) Hum. Mutat. 9, 209–225 [DOI] [PubMed] [Google Scholar]
- 28.Oji Y., Suzuki T., Nakano Y., Maruno M., Nakatsuka S., Jomgeow T., Abeno S., Tatsumi N., Yokota A., Aoyagi S., Nakazawa T., Ito K., Kanato K., Shirakata T., Nishida S., Hosen N., Kawakami M., Tsuboi A., Oka Y., Aozasa K., Yoshimine T., Sugiyama H. (2004) Cancer Sci. 95, 822–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loeb D. M., Evron E., Patel C. B., Sharma P. M., Niranjan B., Buluwela L., Weitzman S. A., Korz D., Sukumar S. (2001) Cancer Res. 61, 921–925 [PubMed] [Google Scholar]
- 30.Miwa H., Beran M., Saunders G. F. (1992) Leukemia 6, 405–409 [PubMed] [Google Scholar]
- 31.Grubb G. R., Yun K., Williams B. R., Eccles M. R., Reeve A. E. (1994) Lab. Invest. 71, 472–479 [PubMed] [Google Scholar]
- 32.Ghanem M. A., Van der Kwast T. H., Den Hollander J. C., Sudaryo M. K., Oomen M. H., Noordzij M. A., Van den Heuvel M. M., Nassef S. M., Nijman R. M., Van Steenbrugge G. J. (2000) Clin. Cancer Res. 6, 4265–4271 [PubMed] [Google Scholar]
- 33.Sukhatme V. P., Cao X., Chang L. C., Tsai-Morris C. H., Stamenkovich D., Ferreira P. C., Cohen D. R., Edwards S. A., Shows T. B., Curran T., Le Beau M. M., Adamson E. D. (1988) Cell 53, 37–43 [DOI] [PubMed] [Google Scholar]
- 34.Dey B. R., Sukhatme V. P., Roberts A. B., Sporn M. B., Rauscher F. J., 3rd, Kim S. J. (1994) Mol. Endocrinol. 8, 595–602 [DOI] [PubMed] [Google Scholar]
- 35.Baron V., De Gregorio G., Krones-Herzig A., Virolle T., Calogero A., Urcis R., Mercola D. (2003) Oncogene 22, 4194–4204 [DOI] [PubMed] [Google Scholar]
- 36.Ahn B. H., Park M. H., Lee Y. H., Min do S. (2007) FEBS Lett. 581, 5940–5944 [DOI] [PubMed] [Google Scholar]
- 37.Krones-Herzig A., Mittal S., Yule K., Liang H., English C., Urcis R., Soni T., Adamson E. D., Mercola D. (2005) Cancer Res. 65, 5133–5143 [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y., Mancarella S., Wang Y., Yue C., Ritchie M., Gill D. L., Soboloff J. (2009) J. Biol. Chem. 284, 19164–19168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma H. T., Patterson R. L., van Rossum D. B., Birnbaumer L., Mikoshiba K., Gill D. L. (2000) Science 287, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 40.Soboloff J., Spassova M., Xu W., He L. P., Cuesta N., Gill D. L. (2005) J. Biol. Chem. 280, 39786–39794 [DOI] [PubMed] [Google Scholar]
- 41.Graham C., Tucker C., Creech J., Favours E., Billups C. A., Liu T., Fouladi M., Freeman B. B., 3rd, Stewart C. F., Houghton P. J. (2006) Clin. Cancer Res. 12, 223–234 [DOI] [PubMed] [Google Scholar]
- 42.Morton C. L., Favours E. G., Mercer K. S., Boltz C. R., Crumpton J. C., Tucker C., Billups C. A., Houghton P. J. (2007) Invest. New Drugs 25, 285–295 [DOI] [PubMed] [Google Scholar]
- 43.Hamilton T. B., Borel F., Romaniuk P. J. (1998) Biochemistry 37, 2051–2058 [DOI] [PubMed] [Google Scholar]
- 44.Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., Housman D., Jaenisch R. (1993) Cell 74, 679–691 [DOI] [PubMed] [Google Scholar]
- 45.Lee S. L., Tourtellotte L. C., Wesselschmidt R. L., Milbrandt J. (1995) J. Biol. Chem. 270, 9971–9977 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.