FIGURE 4.
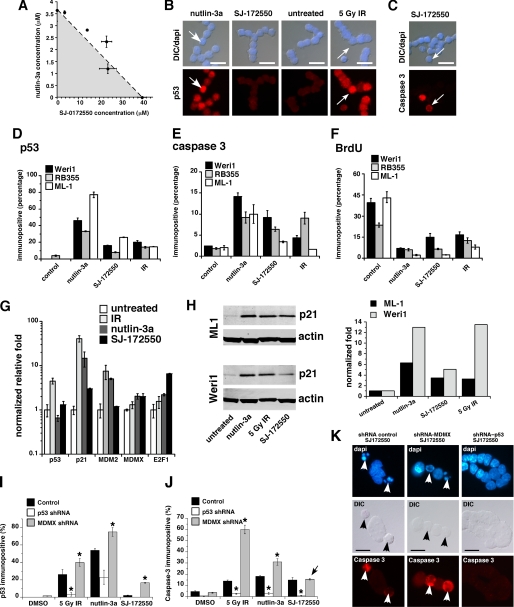
SJ-172550 disrupts the MDMX-p53 interaction in cells maintained in culture. A, isobologram shows the additive inhibition of Weri1 cell growth when the combination of SJ-172550 and nutlin-3a was used to treat cells. Multiple ratios were tested, and the plot shows that the LC50 value (dashed line) was additive but not synergistic (shaded area) or adverse (area above the dashed line). The error bars represent standard deviation from two independent experiments. B and C, immunostaining of compound-treated Weri1 cells. The top panels are merged images of differential interference contrast (DIC) and 4′,6-diamidino-2-phenylindole (DAPI) staining, and the lower panels show p53 levels (B) or caspase-3 activation (C). Nutlin-3a and IR were used as positive controls, and DMSO was used as a negative control. D–F, quantification of the percentage of immunopositive cells shown in B and C and bromodeoxyuridine (BrdU). G, real time PCR quantification of p53 target genes p21, MDM2, and E2F1 activated by drug or IR treatments. MDMX levels were also tested. H, immunoblot and quantification for p21 protein levels activated by drug or IR treatment. Gy, gray. I and J, histogram of the proportion of p53 or activated caspase-3 immunopositive cells following treatment with 5 gray IR, nutlin-3a or SJ-172550. In parallel samples, p53 was knocked down using a p53 shRNA, and MDMX was knocked down using an MDMX shRNA (22), or the samples were treated with a control (scrambled) shRNA. Each bar represents the mean ± S.D. scoring in triplicate of 100 cells for each condition and each shRNA. K, representative images of Weri1 retinoblastoma cells following treatment with SJ-172550 when they lacked p53 (p53 shRNA) or MDMX (MDMX shRNA) as compared with a control shRNA. Arrows indicate activated caspase-3 immunopositive cells in the red (Cy3) channel. shRNA, small hairpin RNA. Scale bars, 10 μm.