FIGURE 6.
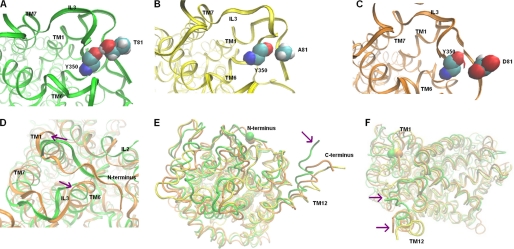
Molecular dynamics simulations of SERTThr-81 mutants reveal models favoring inward facing states. A, snapshot of wild type SERT after 16 ns of MD simulation. The Thr81 side chain forms a stable H-bond with the backbone carbonyl of Tyr350 in IL3. B, snapshot of SERTT81A after 6 ns of MD simulation; the H-bond is not formed between Ala81 and Tyr350 during the course of the simulation. C, snapshot of SERTT81D after 6 ns of MD simulation; no H-bond is formed between Asp81 and Tyr350 during the course of the simulation. D, snapshots of wild type SERT and SERTT81D, following an MD simulation (backbones superimposed). In the in silico mutant (displayed in orange), the cytoplasmic region of TM1 is moved apart from TMD6 and IL3 (indicated by magenta arrows) in comparison with wild type SERT (green). E and F, snapshots of wild type SERT and SERTT81D during MD production simulation (backbones superimposed); TM12 moves away from TM1 (N terminus) after mutation of Thr81 to alanine or aspartate (wild type SERT, green; SERTT81A, yellow; SERTT81D, orange; Cα atoms of Thr81 are displayed in sphere representation).