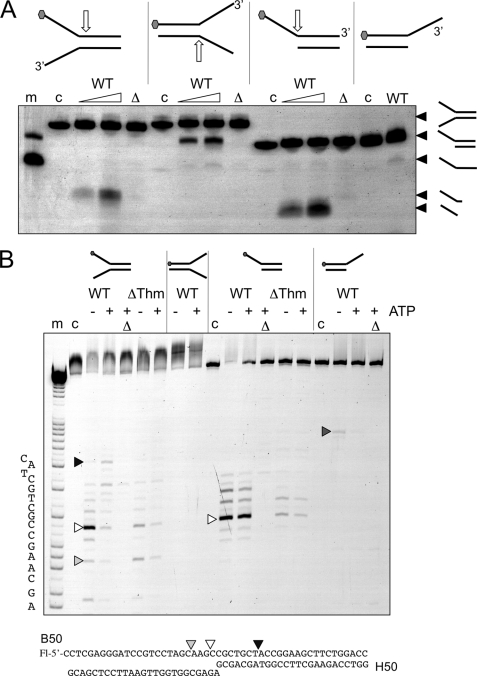
FIGURE 6.
Cleavage of minimal substrates by XPB-Bax1. A, native gel electrophoresis showing cleavage of splayed duplex and overhang substrates. Oligonucleotide B50 with a 5′-fluorescein (gray hexagon) was annealed with a range of partner strands to generate minimal substrates. Approximate cleavage sites are indicated by white arrows. Cleavage products are indicated on the right of the gel. The wild type enzyme (WT) was incubated with each substrate for 5 or 20 min in the presence of ATP and MnCl2. The Δnuc mutant was incubated in the same buffer for 20 min. Control lanes are as follows. m, marker DNA showing migration of overhang and single-stranded B50 oligonucleotide; c, DNA substrate control; Δ, Δnuc mutant. B, the reaction products were also run on denaturing acrylamide TBE gels with (A + G) markers to map cleavage sites. All reactions were carried out in reaction buffer with 10 mm MnCl2 and ATP where indicated. m, A + G sequence markers; c, DNA alone; Δ, Δnuc mutant.