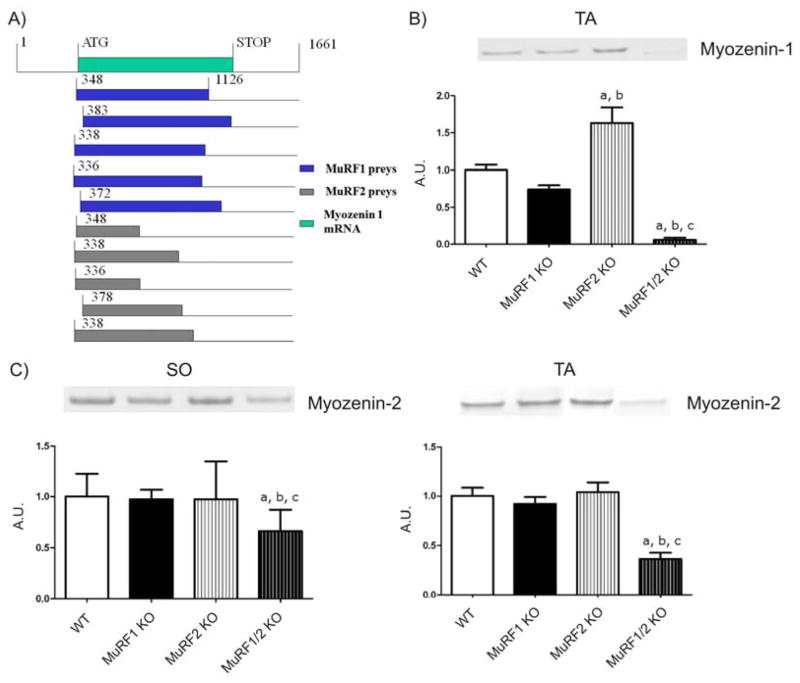
Figure 4. Physical and functional interaction of MuRF1 and MuRF2 with the calsarcin members myozenin-1 and 2.
A) Top green bar shows the translated region of the myozenin-1 cDNA (spanning from bp 348 to 1126; accession NM021245). A total of ten myozenin-1 (MYOZ-1) prey clones were identified in the MuRF1 screen (blue), or in the MuRF2 screen (grey). All ten MYOZ1 prey clones represent full-length or close to full-length cDNA clones. This predicts that the region encompassing nucleotides 383 up to the 3′ end is required for the interaction of MYOZ1 with MuRF1 and MuRF2.
B) Western blot analysis of MYOZ-1 expression in MuRF1, MuRF2 and dKO mice. Consistent with Frey et al (2008) we detected MYOZ1 in tibialis anterior (TA) but not soleus (SO) by Western blots. Combined deletion of MuRF1 and MuRF2 results in loss of MYOZ1 expression in TA. a: p<0.05 vs WT group; b: p<0.05 vs MuRF1 KO group and c: p<0.05 vs MuRF2 KO group; A.U. arbitrary units.
C) Myozenin-2 (MYOZ-2) expression was also reduced after MuRF1/MuRF2 deletion, although the effect was less striking (left: SO; right: TA). a: p<0.05 vs WT group; b: p<0.05 vs MuRF1 KO group and c: p<0.05 vs MuRF2 KO group.