Abstract
Along with silencing intended target genes, transfected siRNAs regulate numerous unintended transcripts through a mechanism in which the equivalent of a microRNA-like seed region in the siRNA recognizes complementary sequences in transcript 3′ UTRs. Amelioration of this off-target silencing would lead to more accurate interpretation of RNA interference (RNAi) experiments and thus greatly enhance their value. We tested whether lentivirus-mediated delivery of shRNA is prone to the sequence-based off-target activity prevalent in siRNA experiments. We compared target gene silencing and overall impact on global gene expression caused by multiple sequences delivered as both transfected siRNAs and lentivirus vector-expressed shRNAs. At equivalent levels of target gene silencing, signatures induced by shRNAs were significantly smaller than those induced by cognate siRNAs and arose less frequently from seed region activity. Interestingly, the low level of seed region-based off-target activity exhibited by shRNAs resulted in down-regulation of transcripts that were largely distinct from those regulated by siRNAs. On the basis of these observations, we recommend lentivirus-mediated RNAi for pathway profiling experiments that measure whole genome transcriptional readouts as well as for large-scale screens when resources for extensive follow up are limited.
Keywords: RNA interference, off-target activity, siRNA, shRNA, lentivirus, seed region
INTRODUCTION
RNA interference (RNAi) is used widely for functional genetic analysis and screens, and holds promise for the development of therapies for currently intractable drug targets. However, RNAi is hampered by widespread siRNA-mediated silencing of multiple transcripts in addition to the intended siRNA target (Jackson et al. 2003; Semizarov et al. 2003). This “off-target” activity is mediated by partial transcript complementarity to a 6–8 base stretch within the 5′ end of the siRNA guide strand that acts like a microRNA seed region (Jackson et al. 2003, 2006b; Birmingham et al. 2006). Multiple studies demonstrated that the seed region regulates both transcription and translation of large numbers of transcripts (Lai 2002; Lim et al. 2005). Thus it has been proposed that off-target transcript silencing is a fundamental, and possibly unavoidable, feature of siRNA-mediated gene silencing.
Our preliminary studies suggested that RNAi mediated by lentivirus-delivered shRNA vectors is less prone to off-target activity than are siRNAs. Microarray analysis of HeLa cells transduced with lentiviral vectors encoding two distinct shRNAs targeting PTEN revealed prominent down-regulation of PTEN itself with little change in the expression of other genes (Klinghoffer et al. 2008). A similar analysis showed that transfection of a TP53-targeting siRNA into colon carcinoma cells resulted in greater numbers of up- and down-regulated transcripts than induction of an shRNA encoding the same core sequence (Rao et al. 2009). Finally, whereas several reports highlight the prevalence of off-target activities in sets of hits from large-scale siRNA screens, hit sets from screens with libraries of arrayed lentiviral shRNA vectors have been generally small and enriched for shRNAs targeting expected pathway regulators (Lin et al. 2005; Ma et al. 2006; Klinghoffer et al. 2008, 2009).
The ability to reduce or eliminate off-target activity would significantly facilitate biological interpretation of experiments involving RNAi-induced gene silencing, streamline complex and costly RNAi screen hit validation schemes designed to work around off-target effects, and figure prominently into the development of RNA-based therapeutics. To date, however, it has been difficult to incorporate elements into siRNA design that consistently ameliorate the vast majority of unintended transcript silencing events (Jackson et al. 2006a). To rigorously confirm preliminary suggestions that lentivirus-mediated delivery of shRNA vectors is less prone to off-target activity compared with transfection of siRNAs, we performed a head-to-head comparison of multiple siRNA/LV-shRNA pairs for target silencing and off-target gene regulation by microarray analysis.
RESULTS/DISCUSSION
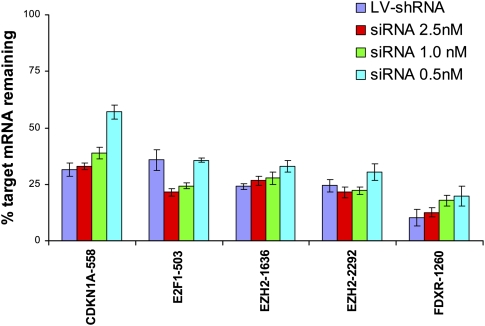
To assess the extent to which lentivirus-mediated RNAi is prone to off-target effects, we compared gene expression profiles induced by siRNAs and shRNAs that encode the exact same 19-mer RNA duplex sequence in HeLa cells. siRNA/shRNA pairs targeting four distinct genes (CDKN1A, E2F1, EZH2, and FDXR) were analyzed. To facilitate the comparison of siRNA and shRNA on- and off-target effects, we identified siRNA concentrations that resulted in levels of silencing similar to that of the corresponding lentivirus-delivered shRNA sequence. Quantitative PCR analysis of target gene silencing yielded transfection concentrations for five siRNA/shRNA pairs (covering all four genes) suitable for our comparison (Fig. 1).
FIGURE 1.
Identification of siRNA concentrations that induce target silencing equal to that induced by corresponding lentiviral shRNA vectors. HeLa cells were either transfected with siRNA by use of RNAiMax at three different concentrations of oligonucleotide duplex or infected with lentiviral vectors encoding the same duplex as a hairpin in the context of the lentiviral vector pLenti6 (LV-shRNA). RNA was harvested 24 h after transfection or 36 h after infection and analyzed by quantitative real-time PCR for target gene silencing by use of gene specific probes from ABI. Error bars, SDs in technical replicates. Target silencing for siRNA transfections (blue bars) was compared with mock transfected cells, whereas silencing induced by LV-shRNA (red bars) was compared with samples infected with the control virus H1-TERM (no shRNA control vector). For gene expression profiling comparisons, siRNA concentrations that resulted in levels of target silencing similar to that induced by LV-shRNA infection were used.
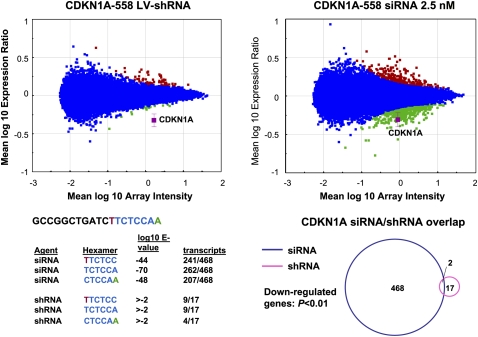
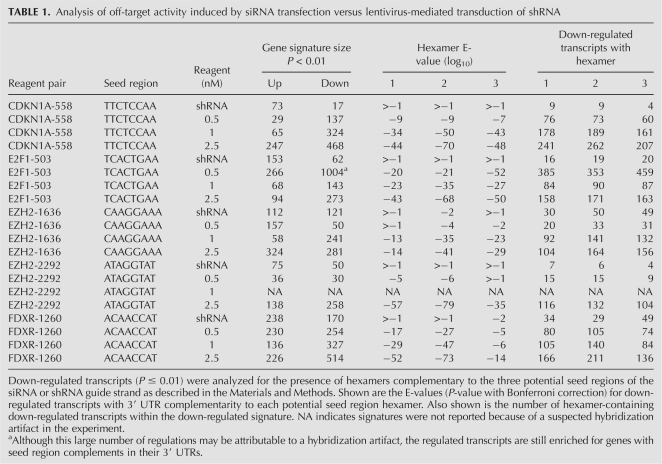
Microarray analysis of each siRNA/shRNA pair revealed multiple differences in the gene expression profiles induced by each method. Consistent with the conclusion that lentivirus-mediated RNAi is not subject to the same off-target activity as the siRNA-induced process, the number of gene expression changes induced by each shRNA was generally smaller compared with the number induced in siRNA profiles (Fig. 2; Table 1). Gene sets down-regulated by both siRNA and shRNA included transcripts containing hexamers complementary to the seed region of the guide strand. However, enrichment for complementary hexamer matches in transcript 3′ UTRs was statistically significant only in the case of the siRNA target gene sets (Fig. 2; Table 1). Interestingly, further analysis of the down-regulated genes revealed very little overlap between the siRNA and shRNA signatures (Fig. 2). This included minimal overlap between seed region-regulated transcripts silenced by siRNA and the corresponding shRNA. Therefore, head-to-head comparison of multiple siRNA/shRNA pairs demonstrated that, whereas both approaches can induce seed region based off-target activity, the extent of this activity driven by shRNA transduction is substantially reduced and different from that mediated by siRNA transfection.
FIGURE 2.
Significant seed region based off-target activity is induced by siRNA but not shRNA delivery conditions that result in equivalent levels of target gene silencing. RNAi was induced in HeLa cells by siRNA transfection or shRNA transduction, RNA was harvested and subjected to microarray analysis. Significance plots show the log10 ratio of gene expression effected by the CDKN1A shRNA compared with the control vector H1-TERM (see Materials and Methods) and the log10 ratio of gene expression by the CDKN1A siRNA compared with mock transfected cells (y-axis) versus the log10 fluorescence intensity of the individual probes on the array (x-axis). Down-regulated transcripts are indicated in green, up-regulated transcripts are shown in red, and the target transcript is shown in magenta. Blue indicates no significant change. Shown is one example of the five paired comparisons in which a duplex introduced as a vector expressed shRNA resulted in a smaller gene signature (P < 0.01) than the same duplex introduced as lipid transfected siRNA. The sense sequence of the 19-mer core is indicated with positions 2–7 of the seed region of the guide strand indicated in blue font. Bases complementary to positions 1 and 8 of the guide strand are indicated in green and red, respectively, to cover the entire seed region of the core sequence. The 3′ UTRs of signature transcripts were surveyed for complementarity to seed region hexamers. Shown are the E-values (P-value with Bonferroni correction) for down-regulated transcripts with 3′ UTR complementarity to each potential seed region hexamer. Also shown is the number of hexamer-containing down-regulated transcripts over the number of total down-regulated transcripts. A Venn diagram of down-regulated transcripts demonstrates that transcripts regulated by the CDKN1A siRNA and the CDKN1A shRNA were largely distinct.
TABLE 1.
Analysis of off-target activity induced by siRNA transfection versus lentivirus-mediated transduction of shRNA
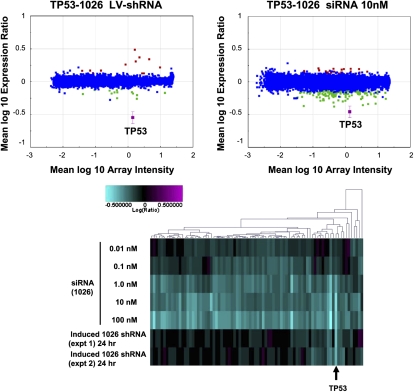
Next, we addressed the possibility that observed differences in off-target activity were attributable to different cellular responses to methods of delivery (transfection of siRNA versus lentivirus transduction of shRNA). Indeed, annotation of up-regulated transcripts highlighted significant virus-mediated immune responses for the majority of the lentivirus-infected samples (E-values ≤10−5; data not shown). This may be due to incomplete normalization of the titers of each shRNA vector with that of the empty vector control (H1TERM). Because a response to infection could impact transcriptional regulation induced by RNAi, we assessed gene expression profiles in cells stably engineered to express an inducible shRNA. For this purpose we generated HCT-116 colon carcinoma cells (wild type for TP53) harboring a stable tetracycline-regulatable expression construct encoding an shRNA designed to silence TP53. Exposure of these cells to the tetracycline analog doxycycline resulted in robust suppression of TP53 with no significant virus response compared with untreated cells (Fig. 3, top; data not shown). We compared transcript regulation induced by doxycycline-mediated expression of a TP53-targeting shRNA to that induced by transfection of the same cells with a TP53 siRNA with the same 19-mer core sequence. Similar to our results with transient infection, induction of the TP53 shRNA resulted in substantially fewer down-regulated genes than transfection of the TP53 siRNA at a concentration that caused a similar level of target silencing (Fig. 3, top). This observation suggested that the smaller number of genes down-regulated by lentivirus-delivered shRNA relative to transfected siRNA was not attributable to an artifact of a virus-mediated immune response. The siRNA titration shown in Figure 3 (bottom) provided evidence that siRNA-induced off-target effects cannot be reduced to the low levels observed with shRNAs simply by decreasing the concentration of siRNA transfected without a corresponding loss in target gene silencing.
FIGURE 3.
An induced TP53 shRNA regulates fewer transcripts than a corresponding siRNA. HCT-116 cells stably expressing a tetracycline regulatable transcriptional repressor and encoding a TP53 shRNA under control of a doxycycline-inducible promoter were treated with doxycycline. Wild-type HCT116 cells were transfected with an siRNA of the same 19-mer core sequence. In the top panels, RNA was harvested 48 h after induction of RNAi and subjected to microarray analysis. Gene expression signatures (P-value <0.01) were determined and plotted as described above. Down-regulated transcripts are indicated in green, up-regulated transcripts are shown in red, and the target transcript is shown in magenta. Blue indicates no significant change. In the bottom panel, RNA from cells treated with the TP53 siRNA or induced with doxycycline to express the TP53 shRNA was harvested 24 h after treatment. For this experiment, we interrogated all ∼22,000 genes on the Agilent array platform used in the experiment. For the cluster analysis, cuts were set such that only genes regulated by at least one experiment in the cluster are shown.
This study represents the most direct evidence that RNAi mediated by lentivirus-delivered shRNA is not subject to the same level of off-target activity revealed by nearly all siRNA-induced gene expression signatures. This difference could be explained by differences in the mechanisms by which lentivirus-encoded shRNAs and lipid-aggregated siRNAs enter the RNAi pathway. Lentiviruses incorporate viral sequences (including an expression cassette for the shRNA) into the host cell genome and require host cell machinery to transcribe and process expressed shRNAs into products that can enter the RNA-induced silencing complex (RISC) to mediate target transcript silencing. It is possible that endogenous processing regulates loading of RISC with internally derived siRNAs and thus decreases off-target activity. Our data are also consistent with several reports that synthetic shRNAs enhance the potency of on-target RNAi-mediated silencing in both in vitro and in vivo models (Kim et al. 2005; Siolas et al. 2005; McAnuff et al. 2007). The increased potency of target gene silencing relative to seed region-based effects mediated by shRNAs may explain why, at equivalent levels of target silencing, microarray analysis revealed fewer hexamer-containing genes down-regulated by shRNA than by siRNA. Moreover, it is conceivable that degradation of unprotected siRNAs in the cytoplasm compromises the integrity of transfected duplexes resulting in more off-target silencing.
We tested a single shRNA structure (a 19-mer stem sequence with a 9 base loop) in a single vector background (pLenti6). Although it is possible that reduced off-target activity is specific to systems similar to the one used in this study, our results show that RNAi with this type of vector-mediated approach could streamline experimental strategies by requiring fewer follow up steps to distinguish on- from off-target effects. This feature of lentivirus-mediated RNAi will be especially important for experiments that make use of global gene expression to profile cellular and disease pathways as well as for high-throughput screens for drug target and biomarker discovery.
MATERIALS AND METHODS
siRNA duplexes and shRNA vectors
All 19-mer core sequences were generated by use of a proprietary algorithm established by Rosetta Inpharmatics scientists. siRNA duplexes were synthesized by Sigma. Vectors were designed to express single-stranded RNA with complementarity that would allow formation of a structure with 19-mer shRNA guide and passenger strands linked by a 9-base hairpin with the following sequence: 5′-TTCAAGAGA-3′. These expression cassettes contained an H1 promoter and an RNA polymerase III termination sequence (TTTTT) flanking the shRNA precursor and were cloned into the lentiviral expression vector pLenti6 (Invitrogen). Sequences used in this study are provided in the supplemental materials (Supplemental Table S1).
Gene silencing by transient transfection and transduction
HeLa cells were transfected in six-well plates with siRNA duplexes at the concentrations indicated by use of the transfection reagent RNAiMax according to the manufacturer's protocols (Invitrogen). shRNAs were cloned into the lentiviral shRNA expression vector pLenti6/V5-DEST (Invitrogen) and were packaged according to the manufacturer's protocols in 293FT cells (Invitrogen) by use of ViraPower Lentiviral Expression reagents (Invitrogen). Packaged lentivirus particles were delivered to cells in the presence of 6 μg/mL polybrene.
Inducible shRNA expression
HCT-116 cells (ATCC) were transduced with lentiviruses expressing tTS (tetracycline-controlled transcriptional repressor; Clontech). Infected cells were selected with G418 as a pooled population. Cells stably expressing tTS were subsequently infected in the presence of 6 μg/mL polybrene with a lentivirus encoding a tetracycline-inducible TP53 shRNA driven from an H1 promoter (Chau et al. 2009). For induction of shRNA expression, cells cultured for periods of greater than 1 wk were incubated in the presence of 50 ng/mL doxycycline, and RNA samples were harvested at the indicated time points following treatment.
Quantitative PCR and microarray analysis
Unless indicated otherwise, RNA was harvested 24 h after siRNA transfection or 36 h after shRNA-expressing lentivirus infection to accommodate a potential lag time between infection and host cell processing of the expressed shRNA into a mature RNA duplex. At earlier time points (12 h for siRNA experiments and 24 h for shRNA experiments), suboptimal silencing was generally observed (data not shown). RNA was extracted from cells by use of RNeasy kits (Qiagen) according to the manufacturer's protocol for animal cells.
mRNA silencing was quantified by real-time PCR with an ABI PRISM 7900HT Sequence Detection System and Assays-on-Demand gene expression products (Applied Biosystems) as described previously (Bartz et al. 2006). The following Assays-on-Demand reagents were used in this study: TP53 (Hs00153340_m1), CDKN1A (Hs00355782_ml), E2F1(Hs00153451_ml), EZH2 (Hs01016789_ml), and FDXR (Hs01031618_gl).
Microarray analysis for all siRNA/shRNA comparisons except for analysis of the inducible HCT-116 cells was performed by use of custom Affymetrix arrays. Extracted RNA was quantified by use of RiboGreen RNA Quantitation Reagent (Invitrogen), and its quality was assessed by use of the Agilent RNA 6000 Pico Kit (Agilent) in an Agilent 2100 Bioanalyzer (Agilent). Samples were amplified and labeled by use of the Ovation WB protocol (NuGEN Technologies), according to the manufacturer's instructions. The resulting amplified cDNAs were hybridized to Affymetrix gene expression chips (Human Rosetta Custom Affymetrix 1.0, Affymetrix). Images were analyzed by use of Affymetrix GeneChip Operating Software (GCOS) and processed further to derive sequence-based intensities by use of the RMA algorithm in the Affymetrix Power Tools (available at http://www.affymetrix.com/support/developer/powertools/index.affx). RNA from cells transfected with siRNAs was compared with RNA from mock transfected controls and RNA from cells transduced with shRNA vectors with RNA from cells infected with a previously described control vector that lacks an shRNA sequence between the H1 promoter and two tandem termination sequences (H1-TERM) (Klinghoffer et al. 2008). Samples from HCT-116 cells were analyzed by use of custom Agilent arrays as described previously (Jackson et al. 2003). In experiments with the stably introduced inducible shRNA construct, RNA from cells treated with doxycycline was compared with RNA from untreated cells. To identify RNAi-regulated genes, ratios of transcript abundance in experimental versus control samples were calculated with normalized intensity data. Data shown are signature genes that display a difference in expression level (P < 0.01) relative to control samples. Data were analyzed by use of Rosetta Resolver or MATLAB software. Analysis of hexamer complementarity was performed as described previously (Jackson et al. 2006b).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Patrick Lewis and the Rosetta Gene Expression Laboratory for managing and processing our array profiling experiments. We dedicate this paper to the superb colleagues with whom we have had the privilege to work with at Rosetta Inpharmatics throughout the years.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1977810.
REFERENCES
- Bartz SR, Zhang Z, Burchard J, Imakura M, Martin M, Palmieri A, Needham R, Guo J, Gordon M, Chung N, et al. 2006. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol 26: 9377–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 3: 199–204 [DOI] [PubMed] [Google Scholar]
- Chau BN, Diaz RL, Saunders MA, Cheng C, Chang AN, Warrener P, Bradshaw J, Linsley PS, Cleary MA 2009. Identification of SULF2 as a novel transcriptional target of p53 by use of integrated genomic analyses. Cancer Res 69: 1368–1374 [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. 2006a. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12: 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS 2006b. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12: 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ 2005. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol 23: 222–226 [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Roberts B, Annis J, Frazier J, Lewis P, Linsley PS, Cleary MA 2008. An optimized lentivirus-mediated RNAi screen reveals kinase modulators of kinesin-5 inhibitor sensitivity. Assay Drug Dev Technol 6: 105–119 [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Frazier J, Annis J, Berndt JD, Roberts BS, Arthur WT, Lacson R, Zhang XD, Ferrer M, Moon RT, et al. 2009. A lentivirus-mediated genetic screen identifies dihydrofolate reductase (DHFR) as a modulator of β-catenin/GSK3 signaling. PLoS One 4: e6892. doi: 10.1371/journal.pone.0006892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC 2002. MicroRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30: 363–364 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y 2005. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res 33: 4527–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA 2006. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443: 359–363 [DOI] [PubMed] [Google Scholar]
- McAnuff MA, Rettig GR, Rice KG 2007. Potency of siRNA versus shRNA mediated knockdown in vivo. J Pharm Sci 96: 2922–2930 [DOI] [PubMed] [Google Scholar]
- Rao DD, Senzer N, Cleary MA, Nemunaitis J 2009. Comparative assessment of siRNA and shRNA off target effects: What is slowing clinical development. Cancer Gene Ther 16: 807–809 [DOI] [PubMed] [Google Scholar]
- Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW 2003. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci 100: 6347–6352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA 2005. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol 23: 227–231 [DOI] [PubMed] [Google Scholar]