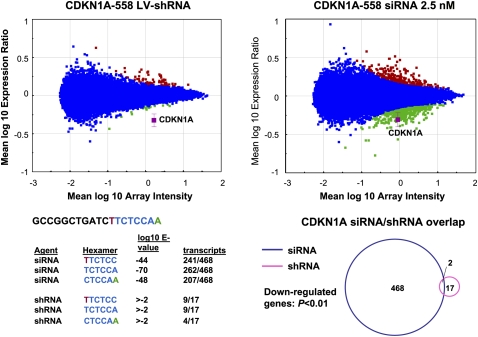
FIGURE 2.
Significant seed region based off-target activity is induced by siRNA but not shRNA delivery conditions that result in equivalent levels of target gene silencing. RNAi was induced in HeLa cells by siRNA transfection or shRNA transduction, RNA was harvested and subjected to microarray analysis. Significance plots show the log10 ratio of gene expression effected by the CDKN1A shRNA compared with the control vector H1-TERM (see Materials and Methods) and the log10 ratio of gene expression by the CDKN1A siRNA compared with mock transfected cells (y-axis) versus the log10 fluorescence intensity of the individual probes on the array (x-axis). Down-regulated transcripts are indicated in green, up-regulated transcripts are shown in red, and the target transcript is shown in magenta. Blue indicates no significant change. Shown is one example of the five paired comparisons in which a duplex introduced as a vector expressed shRNA resulted in a smaller gene signature (P < 0.01) than the same duplex introduced as lipid transfected siRNA. The sense sequence of the 19-mer core is indicated with positions 2–7 of the seed region of the guide strand indicated in blue font. Bases complementary to positions 1 and 8 of the guide strand are indicated in green and red, respectively, to cover the entire seed region of the core sequence. The 3′ UTRs of signature transcripts were surveyed for complementarity to seed region hexamers. Shown are the E-values (P-value with Bonferroni correction) for down-regulated transcripts with 3′ UTR complementarity to each potential seed region hexamer. Also shown is the number of hexamer-containing down-regulated transcripts over the number of total down-regulated transcripts. A Venn diagram of down-regulated transcripts demonstrates that transcripts regulated by the CDKN1A siRNA and the CDKN1A shRNA were largely distinct.