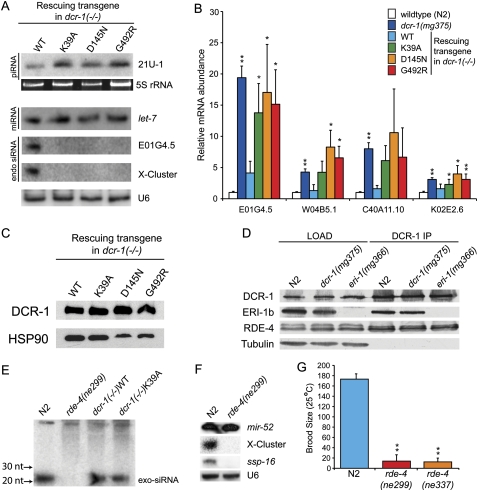
FIGURE 2.
C. elegans with mutations in Dicer's helicase domain are defective for production of endo-siRNAs, but not exo-siRNAs or other small RNAs. (A) Total RNA from mixed stage animals of indicated genotypes was subjected to Northern analysis using probes to a piRNA (21U-1), a miRNA (let-7), or endo-siRNAs (E01G4.5, X-cluster). 5S RNA stained with SYBR Gold, or a second hybridization with U6 snRNA probes, provided loading controls. (B) cDNA from total RNA of indicated strains (colors) was subjected to qRT-PCR to assess mRNA levels for indicated genes (x-axis). mRNA levels were normalized to eft-3 mRNA [N2, dcr-1(mg375); n = 6] or act-1 (transgenic rescue lines; n = 3) and plotted as average ratio of mRNA in mutant animals relative to wild type. Error bars, standard deviation; single asterisks, P < 0.05, and double asterisks, P < 0.01. Identical results were obtained using strand-specific primers/probes for reverse transcription or Northern analysis (data not shown), confirming measured mRNA levels derive from sense rather than antisense transcripts. (C) Embryo extracts of indicated strains were analyzed by Western analysis using α-Flag antibody (top panel) or α-HSP90 antibody (bottom panel) to assess levels of transgenic DCR-1. (D) Embryo extracts of indicated genotype (LOAD) were immunoprecipitated (IP) with an α-DCR-1 antibody. Co-precipitating proteins were analyzed by Western blot using the indicated antibodies. eri-1 encodes two isoforms, and the longer (ERI-1b) interacts with DCR-1 (Duchaine et al. 2006). eri-1(mg366) encodes a premature stop codon, so ERI-1 is not detected in extracts from these animals. Bottom panel, tubulin loading control. (E) Small RNAs were isolated from indicated strains (after feeding E. coli expressing dsRNA to the sel-1 gene), and Northern analyses performed with a probe to detect antisense sel-1. Upper bands are unprocessed sel-1 exo-dsRNA that served as an internal loading control; position of exo-siRNAs is indicated. (F) Small RNAs of mixed stage N2 and rde-4(ne299) C. elegans were subjected to Northern analysis using probes as indicated. Re-probing for U6 served as loading control. (G) N2 and rde-4(ne299) worms (n = 7) were grown at 25°C, and progeny counted to obtain average brood size. Asterisks, P < 0.01.