Figure 5.
Assembly and Sedimentation Analysis of Nucleosome Arrays Bearing Homogeneously Acetylated Nucleosomes
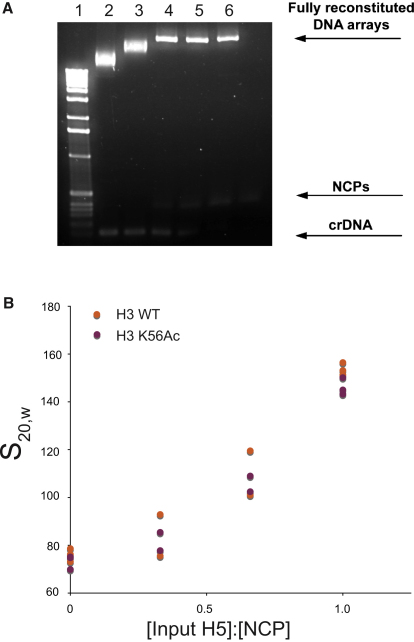
(A) Titration of purified histone H3 K56Ac octamers to assemble chromatin arrays containing 61 repeats of 197 bp of the 601 nucleosome-positioning DNA sequence. A retarded gel shift indicates loading of the DNA array with histone octamers. Excess histone octamer forms nucleosome core particles (NCPs) with competitor DNA (crDNA). Conditions of lane 4 were used to reconstitute DNA arrays in subsequent experiments.
(B) DNA arrays were reconstituted with saturating amounts of histone octamer and with increasing amounts of H5 linker histone in order to induce compaction. Chromatin arrays were folded in 1 mM MgCl2 and 10 mM TEA (pH 7.4), and the degree of the compaction was measured quantitatively by sedimentation velocity analysis.