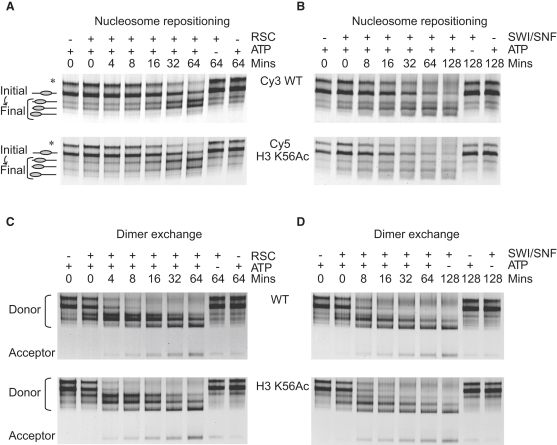
Figure 6.
H3 K56 Acetylated Nucleosomes Cause Minimal Alteration to the Initial Rate of RSC or SWI/SNF Repositioning
(A and B) Competitive repositioning assays were performed using 1 pmol each of H3 K56Ac and wild-type nucleosomes, 1 mM of ATP, and 41 fmol of RSC (A) or 115 fmol of SWI/SNF (B). A representative native PAGE gel of the repositioning assay is shown for each remodeler. The initial rate estimate for repositioning of H3 K56Ac nucleosomes relative to wild-type for RSC was 1.2 fold ± 0.1 (mean ± SE of the mean) and for SWI/SNF was 1.4 fold ± 0.2. Each experiment was repeated in triplicate. Asterisks indicate the P position. WT, wild-type.
(C and D) H3 K56Ac and wild-type nucleosomes exhibit equivalent remodeler-driven dimer transfer. Remodeler dimer transfer was performed using 0.25 pmol of donor nucleosomes assembled with Cy5-labeled H2A onto 54A18 DNA fragments, 0.75 pmol of wild-type tetrasome acceptor assembled on 0W0 DNA fragments, 1 mM of ATP, and 83 fmol of RSC (C) or 230 fmol of SWI/SNF (D). For each dimer transfer experiment, a representative Cy5 scan of the native PAGE gel is shown. Both RSC and SWI/SNF caused a 1.2-fold increase of the percentage of dimer transfer for H3 K56Ac nucleosomes relative to wild-type at the finish of their respective time courses. As the SE of the mean was large in both cases, 0.1 and 0.2 for RSC and SWI/SNF, respectively, there was no significant change in the percentage of dimer transfer. Each experiment was repeated in triplicate.