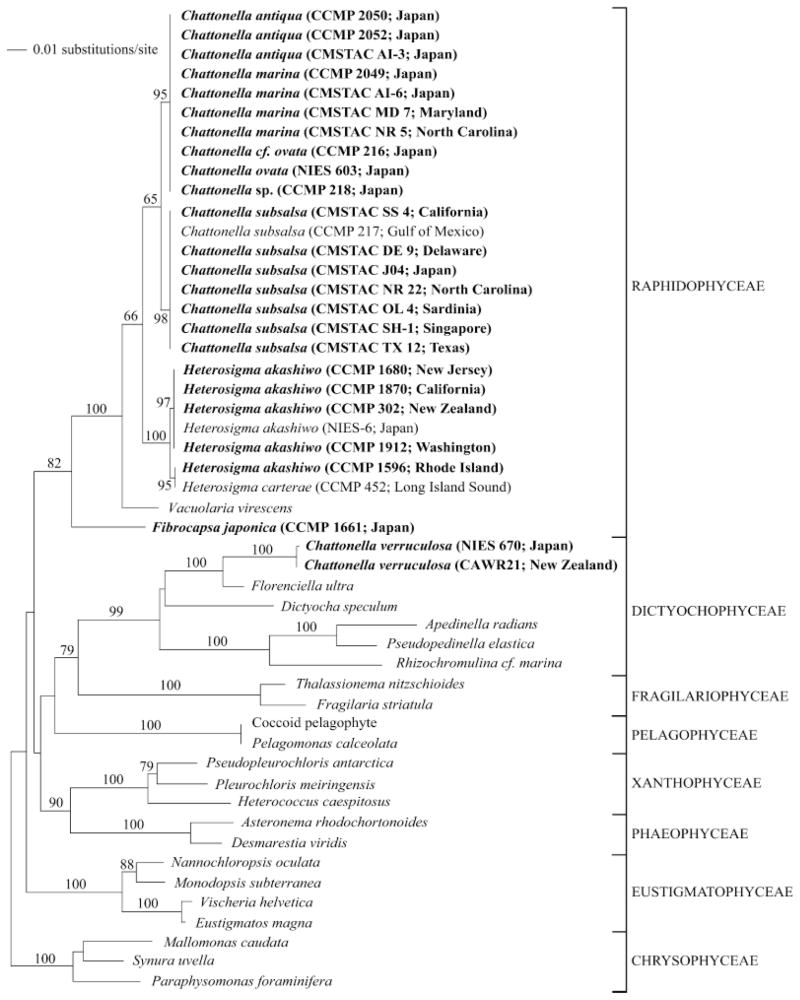
Fig. 1.
Minimum evolution analysis of the 18S rRNA alignment using maximum likelihood distances (distance score: 1.30857). Bootstrap values above 60% are indicated. The following GenBank accession numbers, not listed in Table 1, were included in the phylogenetic analyses: Chattonella subsalsa U41649; Vacuolaria virescens U41651; Heterosigma akashiwo AB001287; H. carterae U41650; Apedinella radians U14384; Pseudopedinella elastica U14387; Rhizochromulina cf marina U14388; Dictyocha speculum U14385; Florenciella ultra AY254857; Coccoid pelagophyte U40927; Pelagomonas calceolata U14389; Pleurochloris meiringensis AF109728; Pseudopleurochloris antarctica AF109729; Heterococcus caespitosus AF083399; Asteronema rhodochortonoides AB056156; Desmarestia viridis AJ295828; Fragilaria striatula X77704; Thalassionema nitzschioides X77702; Eustigmatos magna U41051; Vischeria helvetica AF045051; Monodopsis subterranea U41054; Nannochloropsis oculata AF045044; Mallomonas caudata U73228; Paraphysomonas foraminifera AF174376; and Synura uvella U73222. Cultures in bold were deposited as part of this study, and collapsed branches indicate identical sequences.