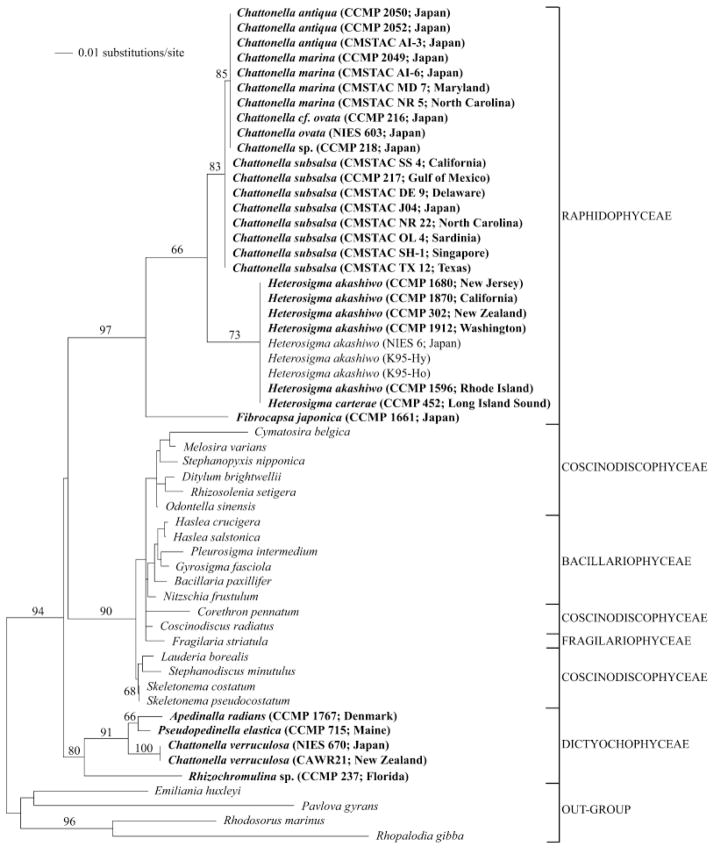
Fig 2.
Minimum evolution analysis of the 16S rRNA alignment using maximum likelihood distances (distance score: 1.05415). Bootstrap values above 60% are indicated. The following GenBank accession numbers, not listed in Table 1, were included in the phylogenetic analyses: Heterosigma akashiwo AB181955, AB181956, AB181958; Cymatosira belgica AJ536456; Melosira varians AJ536464; Stephanopyxis nipponica AJ536465; Ditylum brightwellii AJ536460; Rhizosolenia setigera M87329; Odontella sinensis AJ536457; Haslea crucigera AF514849; Pleurosigma intermedium AF514848; Haslea salstonica AF514854; Bacillaria paxillifer AJ536452; Corethron pennatum AJ536466; Fragilaria striatula AJ536453; Gyrosigma fasciola AF514847; Nitzschia frustulum AY221721; Coscinodiscus radiatus AJ536462; Lauderia borealis AJ536459; Stephanodiscus minutulus AY221720; Skeletonema costatum X82154; Skeletonema pseudocostatum X82155; Rhodosorus marinus AF170719; Rhopalodia gibba AJ582391; Emiliania huxleyi X82156; and Pavlova gyrans AF172715. Cultures in bold were deposited as part of this study, and collapsed branches indicate identical sequences. Because this analysis is based on short sequences (approximately 640 bp), it is shown to depict sequence diversity and should not be used to draw taxonomic conclusions.