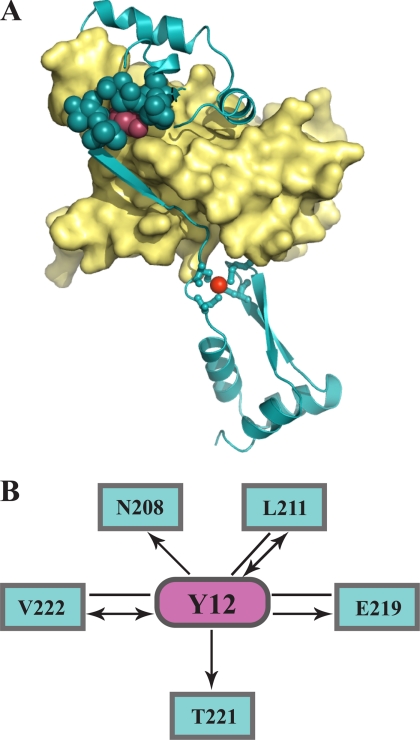
FIGURE 5.
Tyrosine 12: a central interaction hub in Hsp33. A, homology modeling of reduced E. coli Hsp33 was conducted with MODELLER using the crystal structure of reduced B. subtilis Hsp33 (PDB code: 1VZY) as a template. The N-terminal core domain is shown in the yellow surface representation, and the C-terminal redox switch domain is shown in cyan ribbon presentation. The zinc ion is depicted as red sphere. The N-terminal Tyr12 (magenta spheres) appears to interact tightly with several linker residues (cyan spheres). A high GA341 score of 1 provided by the MODWEB modeling server indicates that the model is accurate. B, connectivity map of Tyr12. Interactions between Tyr12 and residues in the Hsp33 C terminus (amino acids 179–288) are depicted as magenta and cyan nodes, respectively. The interaction types shown are van der Waals interactions. Arrows point toward the residues that contribute a backbone atom to the interaction.