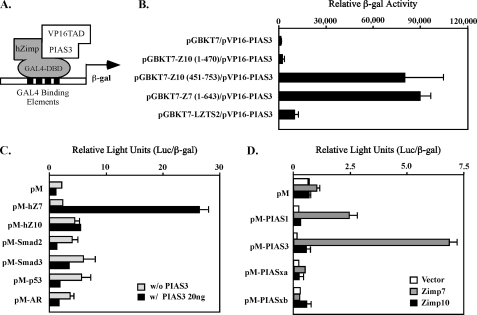
FIGURE 1.
Specific interaction between Zimp7 and PIAS3 proteins. A, a schematic representation of the yeast two-hybrid assay for demonstrating the interaction between PIAS3 and hZimp proteins. B, the pVP16-PIAS3 plasmid containing VP16-TAD and PIAS3 (amino acid 365–482) was co-transformed with pGBKT7 vector or different pGBKT7 fusion constructs. Numbers correspond to amino acid residues. Transformed cells were plated on SD-Ade-Leu-Trp plates and SD-Leu-Trp plates to monitor transformation efficiency. Three independent colonies were inoculated from each transformation experiment for subsequent liquid β-galactosidase assays. The data for the liquid β-galactosidase assays are shown as the mean ± S.D. C, the full-length human Zimp7, Zimp10, AR, SMAD2, SMAD3 or p53 were fused to Gal4 DBD in the pM vector. CV-1 cells were cotransfected with the pM constructs with or without a PIAS3 expression construct, a luciferase reporter construct containing Gal4 binding sites within the chicken myelomonocytic growth factor gene minimal promoter (−41 to +61), and a constitutive β-galactosidase reporter. Data are presented in the RLUs, which were obtained by normalizing the activities of luciferase to those of β-galactosidase. Individual transfection experiments were done in triplicate, and the results reported as the mean ± S.D. D, the full-length PIAS1, PIAS3, PIASxα, or PIASxβ were fused to pM vector, and co-transfected into CV-1 cells with full-length hZimp7 or Zimp10 proteins. The luciferase assay was performed as described above.