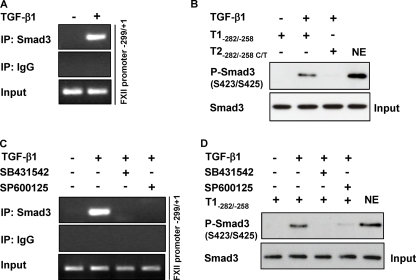
FIGURE 8.
Smad3-SBE-(−272/−269) interaction is suppressed in the presence of a JNK inhibitor. A, HLF were either not stimulated or were stimulated with TGF-β1, and ChIP analysis was performed using a Smad3 antibody or IgG isotype control. PCR was performed with immunoprecipitated (IP) DNA as described under “Experimental Procedures,” thereby producing an amplicon comprising the −299/+1 region of the FXII promoter. PCR products were separated by agarose gel electrophoresis and detected by staining with ethidium bromide. Data are representative of three independent experiments. B, nuclear extracts (NE) from untreated or TGF-β1-treated HLF were incubated with biotinylated templates (T1−282/−258 or T2−282/−258 C/T), and bound Smad3 was detected by Western blotting. Data are representative of three independent experiments. Nuclear extract was used as positive control. C, HLF were pretreated with SB431542 or SP600125 for 1 h prior to incubation with TGF-β1. ChIP analysis was performed using Smad3 antibody or isotype IgG control. PCR was performed with immunoprecipitated chromatin as described under “Experimental Procedures,” thereby producing an amplicon comprising the −299/+1 region of the FXII promoter. Data are representative of three independent experiments. D, HLF were preincubated with SB431542 or SP600125 for 1 h prior to addition of TGF-β1. Nuclear extracts were prepared and then incubated with the biotinylated template T1-(−282/−258). Smad3 was detected by Western blotting. Data are representative of three independent experiments. Nuclear extract was used as positive control.