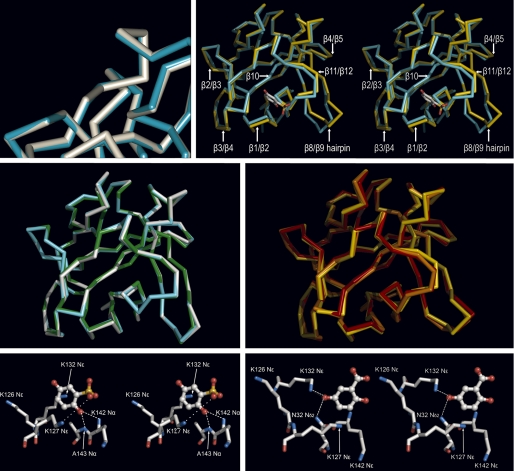
FIGURE 3.
Binding of GA, 2,5DHPS, heparin, and 5A2NMS to FGF-1. Top row, left, shown is a superimposed three-dimensional structure of the FGF-1 backbone bound to 2,5DHPS (cyan) and GA (white) at the inhibitor binding site represented in Fig. 2; right, shown is a stereoview of the superimposed three-dimensional structure of the FGF-1 backbone bound to 2,5DHPS (cyan) and heparin (yellow) (45). For orientation purposes 2,5DHPS is represented at its binding site as a stick model, with the carbon, oxygen, and sulfur atoms colored white, red, and yellow, respectively. Representations were generated with the Discovery Studio Visualizer program in the first case and with the PyMOL program in the latter. In both cases the backbones of the superimposed molecules were reciprocally oriented to a minimal root mean square deviation with the program used to generate their representations. Displacement of the backbone at the level of the β3/β4 loop is not properly appreciated at the figure because the perspective of the drawing. Middle row, left, shown are superimposed three-dimensional structures of the amide backbone of FGF-1 bound to 2,5DHPS (cyan), GA (white), and 5A2NMS (green). Middle row, right, shown are superimposed three-dimensional structures of the amide backbone of FGF-1 bound to heparin (yellow) and incorporated into two different models of the FGF-1·FGFR2 complexes (orange, symmetric; red, asymmetric). The complex also includes heparin in the case of the asymmetric model (45, 51, 57). Superimposed molecules were reciprocally oriented to a minimal root mean square deviation and represented using PyMOL. Bottom row, shown is a network of non-covalent interactions between FGF-1 and 2,5DHPS (left) and GA (right). The ligands and the protein are shown as stick-and-ball models, respectively, with the atoms colored as above. Superimposed molecules were reciprocally oriented to a minimal root mean square deviation and represented using PyMOL.