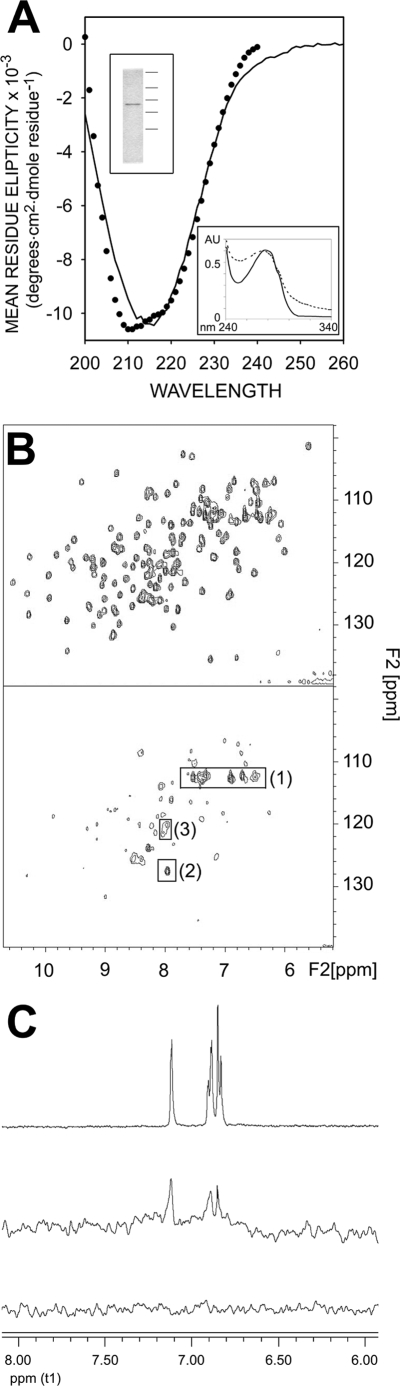
FIGURE 7.
Characterization of recombinant exdFGFR2IIIc. A, circular dichroism spectra in the far-UV region of exdFGFR2IIIc are shown. Closed circles represent the spectrum reconstructed on the basis of the percentage of the secondary structure components of exdFGFR2IIIc obtained by deconvolution of the experimental spectrum (DICHROWEB server (68, 90). Upper inset, Coomassie Brilliant Blue-stained SDS/polyacrylamide electrophoresis gel (15%) (50) of recombinant exdFGFR2IIIc after purification and refolding (the horizontal lines to the right indicate the position of molecular mass markers of 250, 75, 50, 37, and 25 kDa). Lower insert, UV spectra (∼0.5 mg/ml; 20 mm Hepes, 150 mm NaCl (pH 7.5)) of exdFGFR2IIIc before (dotted line) and after refolding are shown. AU, absorbance units. B, shown is a characteristic 1H,15N HSQC spectrum of the backbone of 15N-labeled FGF-1 (150 μm in 20 mm sodium phosphate (pH 7.2), 300 mm NaCl) in the presence (lower panel) and absence (upper panel) of 50 μm exdFGFR2IIIc. Framed 1H,15N cross-peaks correspond to Gln-54 and Gln-91 Nδ and His-138 Nα (1), Asp-154 Nα (2), and Lys-23 and Lys-24 Nα (3). C, shown is an STD spectrum of a solution of 2,5DHPS and exdFGFR2IIIc in 20 mm sodium phosphate, 150 mm NaCl (pH 7.2) at a molar ratio of 1 to 0.05 (middle spectrum). The top record is a 1H NMR spectrum of the aromatic region of a 1 mm solution of 2,5DHPS in the same buffer. The bottom trace is a STD spectrum equivalent to that of the middle trace, except that the protein was omitted. Circular dichroism spectra in the far UV region were obtained using a Jasco 710 spectropolarimeter at 20 °C in a 1-mm path length cuvette. The protein (0.13 mg/ml) was in 50 mm sodium phosphate (pH 7.0), and the spectra were averaged by accumulating four scans. A four-scan averaged spectrum of the buffer was routinely subtracted from the protein spectra.