FIGURE 9.
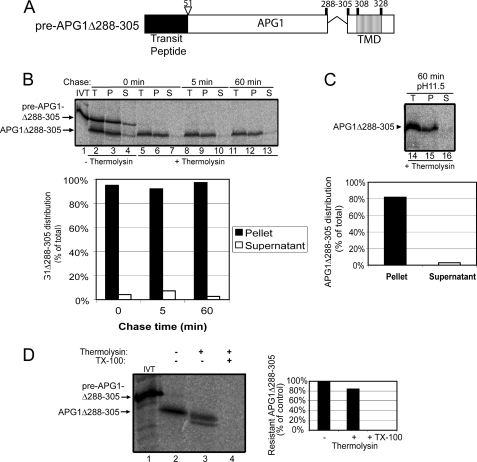
The 20 amino acids upstream of the APG1 TMD do not influence IEM targeting or membrane topology. A, schematic of the pre-APG1Δ288–305 protein. B, [35S]pre-APG1Δ288–305 was imported into chloroplasts for 5 min at 20 °C. The reaction was stopped on ice and treated with thermolysin, and import was resumed in the presence of 5 mm ATP (Chase) for the times indicated. Equivalent fractions were collected and separated into membrane (P) and supernatant (S) fractions by osmotic lysis. The graph represents the quantification of the distribution of APG1Δ288–305 during the chase. C, samples from the 60 min time point in B were treated with 0.2 m Na2CO3, pH 11.5, and separated into soluble and membrane fractions. Lane 14 (T) contains a sample equivalent to the starting material before alkaline treatment. The graph represents the distribution of APG1Δ288–305 between the membrane pellet and supernatant fractions. D, [35S]pre-APG1Δ288–305 was imported into isolated chloroplasts for 30 min at 26 °C and subsequently treated with 200 μg/μl thermolysin for 30 min on ice. The chloroplasts were lysed, and inside-out IEM vesicles were isolated by gradient density centrifugation. The vesicles were treated in the presence (+) or absence (−) of 20 μg of thermolysin/mg of protein in the presence (+) or absence (−) of 2% Triton X-100 (TX-100) as indicated. The graph represents the quantification of the protease-resistant imported protein as indicated.