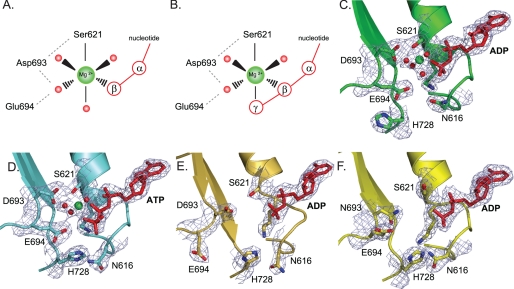
FIGURE 1.
Structural details of magnesium binding sites in wild type MutS and mutant D693N. A and B, schematic view of octahedral magnesium coordination by MutS bound to ADP (Protein Data Bank accession number 1E3M (4)) (A) and ATP (Protein Data Bank accession number 1W7A (13)) (B). The metal ion is coordinated by six ligands: the side chain hydroxyl group of Ser-621, the β-phosphate, and four ordered water molecules. In the case of ATP, the γ-phosphate has displaced one of the ordered water molecules. Asp-693 hydrogen bonds with Ser-621 and coordinates one of the ordered waters; the catalytic base, Glu-694, is in close proximity to this same water molecule. An extensive hydrogen bonding network is formed among the oxygens from the Walker B carboxylates, Ser-621, the nucleoside phosphate groups, and the hydration shell waters. C, magnesium coordination in the A subunit of wild type MutS bound to ADP (1E3M). D, magnesium coordination in the A subunit of wild type MutS bound to ATP (1W7A) and rotations of the His-728 and Asn-616 side chains. E, absence of magnesium and reordering of the Walker B motif in the 16 AA MutS crystal structure. F, absence of magnesium in the crystal structure of D693N MutS.