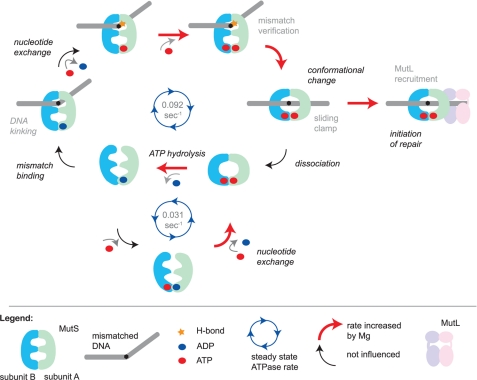
FIGURE 6.
Model for initiation of DNA mismatch repair and the role of magnesium in specific reaction steps. MutS slowly hydrolyzes ATP when it is not bound to DNA. The rate-limiting step in this reaction is the release of ADP from the high affinity nucleotide binding site. The asymmetric ATPase sites are tightly coupled in a manner that depends on correct magnesium coordination by the Walker B motifs. When MutS recognizes a mismatch, it stably binds by kinking the DNA and forming a hydrogen bond between a conserved glutamate and one of the mismatched bases. This results in an increase in the release of ADP from the high affinity site, which is independent of magnesium. However, in the next step, the affinity for ATP strongly depends on magnesium. MutS then uses the hydrogen bond formed between Glu-38 and one of the mismatched bases to verify mismatch binding and undergo an ATP-dependent conformational change into a sliding clamp. If magnesium is not coordinated correctly, the sliding clamp can be formed but at a much reduced rate. Sliding clamps either recruit MutL and thereby initiate the repair reaction or, when idle, will ultimately dissociate from the DNA upon or followed by ATP hydrolysis. MutS is then reset into a conformation that is again able to search for a DNA mismatch.