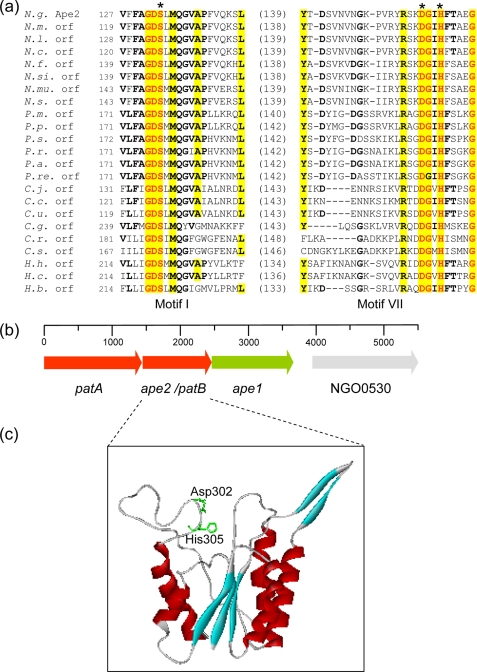
FIGURE 2.
Sequence, genetic organization, and structure of Ape2. a, partial sequence alignment of hypothetical family 2 Ape involving consensus motifs I and VII. The residues in bold face and yellow highlight denote greater than 50 and 80% identity, respectively, while the residues in red are invariant within the family. The values in parentheses denote the number of residues between the respective partial sequences, and the asterisks denote the putative catalytic residues. Abbreviations (accession numbers): N.g., N. gonorrhoeae Ape2 (YP_207683); N.m., N. menningitidis (NP_284201); N.l., N. lactamica (ZP_03722920); N.c., N. cinerea (ZP_03744458); N.f., N. flavescens (EEG32498); N.s., N. subflava (ZP_03750678); N.mu., N. mucosa (ACDX02000005.1); N.si., N. sicca (EET44325); P.m., P. mirabilis (YP_002152370); P.p., P. penneri (EEG87531); P.s., Providencia stuartii (ZP_02961974); P.r., Prov. rustigianii (ZP_03315886); P.a., Prov. alcalifaciens (EEB46552); P.re., Prov. rettgeri (EEF72588); C.j., Campylobacter jejuni (NP281793); C.c., C. coli (ZP_00366779); C.u., C. upsaliensis (ZP_00369961); C.g., C. gracilis (EEV17613); C.r., C. rectus (EEF14732); C.s., C. showae (EET79176); H.h., Helicobacter hepaticus (NP860614); H.c., H. cinaedi (ABQT01000013); H.b., H. bilis (ACDN01000074). b, genetic organization of the poa cluster on the N. gonorrhoeae chromosome including the gene encoding NGO0530, a hypothetical acyl-CoA synthetase. c, three-dimensional structure of the N. gonorrhoeae Ape2 as predicted using PHYRE involving residues Asp-125—Pro-326. The Ape2 residues were threaded onto the SGNH hydrolase thioesterase I (GenBankTM Acc. No. D1jrla) with 17% identity and an E-value of 1.6−19. The position of the putative catalytic Asp-302 and His-305 residues are illustrated within a groove through the middle of the enzyme; the N-terminal segment containing Ser-133 was not modeled by the algorithm.