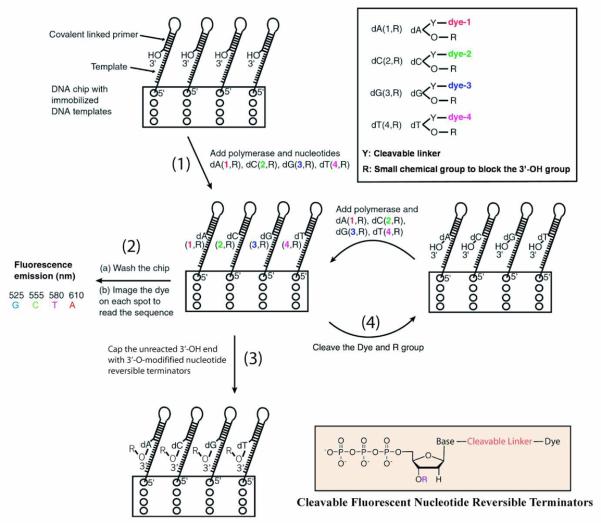
Figure 2.
In the SBS approach, a chip is constructed with immobilized DNA templates that are able to self-primefor initiating the polymerase reaction. Four nucleotide analogues are designed such that each is labeled with aunique fluorescent dye on the specific location of the base, and a small chemical group (R) to cap the 3′-OH group. Upon adding the four nucleotide analogues and DNA polymerase, only the nucleotide analogue complementary tothe next nucleotide on the template is incorporated by polymerase on each spot of the chip (step 1). After removingthe excess reagents and washing away any unincorporated nucleotide analogues, a 4 color fluorescence imager isused to image the surface of the chip, and the unique fluorescence emission from the specific dye on the nucleotideanalogues on each spot of the chip will yield the identity of the nucleotide (step 2). After imaging, the small amountof unreacted 3′-OH group on the self-primed template moiety will be capped by excess 3′-O-modified nucleotidereversible terminators and DNA polymerase to avoid interference with the next round of synthesis forsynchronization (step 3). The dye moiety and the R protecting group will be removed to generate a free 3′-OH groupwith high yield (step 4). The self-primed DNA moiety on the chip at this stage is ready for the next cycle of thereaction by repetition of steps 1, 2, 3, and 4 to identify the next nucleotide sequence of the template DNA.