Abstract
All canonical transfer RNAs (tRNAs) have a uridine at position 8, involved in maintaining tRNA tertiary structure. However, the hyperthermophilic archaeon Methanopyrus kandleri harbors 30 (out of 34) tRNA genes with cytidine at position 8. Here, we demonstrate C-to-U editing at this location in the tRNA’s tertiary core, and present the crystal structure of a tRNA-specific cytidine deaminase, CDAT8, which has the cytidine deaminase domain linked to a tRNA-binding THUMP domain. CDAT8 is specific for C deamination at position 8, requires only the acceptor stem hairpin for activity, and belongs to a unique family within the “cytidine deaminase–like” superfamily. The presence of this C-to-U editing enzyme guarantees the proper folding and functionality of all M. kandleri tRNAs.
The gene sequence of RNA molecules can be altered by editing enzymes that catalyze the deamination of adenosine or cytidine (1). In transfer RNA (tRNA), adenosine deaminases acting on tRNA engage in adenosine-to-inosine (A-to-I) editing (2–4) and are involved in the formation of 1-methylinosine found in some archaeal tRNAs (5). Cytidine-to-uridine (C-to-U) editing in tRNA is seen in plant mitochondrial (6), eukaryotic, organellar (7), and cytoplasmic Trypanosoma brucei tRNA (8), but the enzyme(s) involved in these conversions are unknown. In Methanopyrus kandleri, only four tRNAs( , and ) possess a U8 (T8 in the tRNA gene), whereas the other 30 tRNA genes encode C8 (9, 10). The highly conserved U8 in tRNAs forms a reverse Hoogsteen tertiary base pair with A14 that stabilizes the sharp kink between the acceptor stem and base A9 (fig. S1) (11). A U8C mutation is associated with mitochondrial myopathy in humans (12) and destabilizes the tRNA structure, resulting in a molecule unfit in translational initiation and elongation (13). The C8 must be modified to U in order to support the tertiary interaction with A14, which is present in all 34 tRNAs.
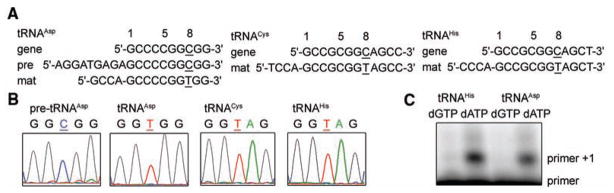
Sequencing of total small RNA from M. kandleri revealed that mature tRNAAsp, tRNACys, and tRNAHis species contain U8, whereas their respective genes have C8 (Fig. 1, A and B). Furthermore, precursor tRNAAsp molecules containing 5′ leader and 3′ trailer sequences still have C8, indicating that C-to-U editing occurs at the RNA level (Fig. 1, A and B). Primer extension of an oligonucleotide annealed to the tRNA directly downstream of base 8 with either 2′-deoxyguanosine 5′-triphosphate (dGTP; complementary to the unedited C8) or 2′-deoxyadenosine 5′-triphosphate (dATP; complementary to the edited U8) revealed that most of base C8 in tRNAAsp and tRNAHis is edited to U (Fig. 1C).
Fig. 1.
C8-to-U8 editing in vivo. (A) Sequencing results for the indicated tRNA gene and reverse transcription polymerase chain reaction (RT-PCR) products of circularized precursor tRNA (pre, with 5′ and 3′ extensions) and mature tRNA (mat). Only the mature tRNA displays the edited U8 (T8 in DNA, underlined). (B) Sequencing traces of RT-PCR products with base 8 underlined. (C) The majority of tRNAs are edited to U8 as minisequencing detects extension of a primer to base 8 in the presence of dATP but not dGTP.
A computational search for C-to-U editing enzyme candidates in M. kandleri used the conserved cytidine deaminase signature zinc-binding motif His-(Ala-Val)-Glu-X24–36-Pro-Cys-X-X-Cys (where X is any amino acid) (14), and yielded one candidate, the orphan protein MK0935, which also contained a THUMP domain, shown to mediate activity on the tertiary core of tRNA in thiouridine and pseudouridine synthases (15, 16), and RNA methylases (15, 16). The MK0935 gene (renamed CDAT8, “cytidine deaminase acting on tRNA base C8”) was cloned, expressed in Escherichia coli, and the recombinant enzyme purified.
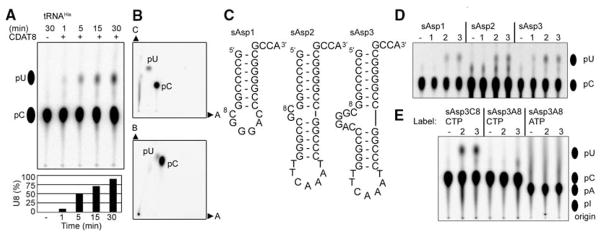
The C-to-U editing activity of CDAT8 was verified in vitro by measuring the deamination of cytidine residues including C8 in M. kandleri tRNAHis transcribed in the presence of [α-32P]cytidine 5′-triphosphate (CTP) (Fig. 2, A and B) (17). To analyze the substrate recognition mechanism of CDAT8, we constructed three minimal tRNA substrates based on M. kandleri tRNAAsp (Fig. 2C). The smallest substrate (sAsp1 in Fig. 2C) consists only of the acceptor stem microhelix with a bulged-out C8G9GGAC sequence, where GGAC represents the variable loop of tRNAAsp. The slightly larger substrates, sAsp2 and sAsp3, additionally contained the tRNA T-stem/loop with bulged-out C8G9 and C8G9GGAC sequences, respectively. All three minimal constructs were substrates for CDAT8, indicating the dispensability of the tRNA anticodon and D-arm for activity (Fig. 2D). Although all three constructs were edited by CDAT8 at 70°C, only sAsp3 was a substrate at 37°C. These results suggest that CDAT8 is able to recognize 30 different tRNAs mainly by interacting with the tRNA acceptor stem, including the universal 3′-CCA end, and a flexible base C8. A modified sAsp3 substrate containing A8 instead of C8 abolished editing activity (Fig. 2E). An A-to-I tRNA deaminase was reported to also perform C-to-U editing on DNA (8). As a test of the possibility of A-to-I editing, the same construct was transcribed with [α-32P]ATP to generate a radioactively labeled A8, which was not edited by CDAT8 (Fig. 2E); this result indicated that CDAT8 performs only C-to-U deamination.
Fig. 2.
C8-to-U8 editing in vitro. (A) Thin-layer chromatography (TLC) analysis of nuclease P1–digested M. kandleri tRNAHis transcripts after reaction with BSA (−) and 10 μM CDAT8 for the indicated times (+) at 70°C. The formation of U8 (C8 is one of 30 labeled cytidine residues in tRNAHis) is quantified in the lower panel. (B) Two-dimensional TLC analysis of the reaction product confirms the identity of the cytidine 5′-monophosphate (pC) and uridine 5′-monophosphate (pU) spots in three different solvents (denoted A, B, and C). The pC and pU markers were visualized by ultraviolet shadowing. (C) Sequence and predicted secondary structures of the three M. kandleri tRNAAsp–derived minimal substrates sAsp1, sAsp2, and sAsp3. (D) TLC analysis of the indicated nuclease P1–digested transcripts after reaction with BSA (−) for 30 min of incubation at 70° and 10 μM CDAT8 after 30 min of incubation at 37°C (1), 50°C (2), or 70°C (3). The additional spots for sAsp2 are generated by nuclease P1 independent of CDAT8 activity. (E) TLC analysis of nuclease P1–digested sAsp3 or its C8A mutant (sAsp3A8) after incubation. The internal label ([α-32P]CTP or [α-32P]ATP) of the transcript is indicated.
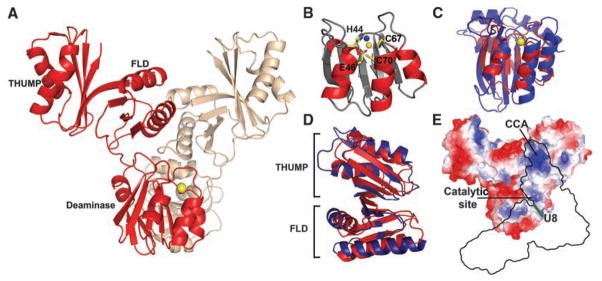
The structure of CDAT8 was solved in two different crystal forms. CDAT8 is composed of three domains: an N-terminal cytidine deaminase domain (CDD), a central ferredoxin-like domain (FLD), and a C-terminal THUMP domain (Fig. 3A). The cytidine deaminase active site has the signature motif His-Ala-Glu-Xn-Pro-Cys-X-X-Cys in which the histidine, two cysteine residues, and a water molecule (Fig. 3B) are involved in the tetrahedral coordination of zinc. The CDD of CDAT8 has the same core structure as those of known cytidine deaminases such as APOBEC2 (18) and APOBEC3G (19, 20) (Fig. 3C). However, the CDAT8 CDD is much more compact, perhaps because its substrate (tRNA) recognition is provided by additional domains. The FLD and THUMP domains are highly homologous to those of ThiI (16) with backbone root mean square deviations of 2.8 and 2.2 Å, respectively (Fig. 3D). These two domains are believed to be responsible for tRNA recognition by ThiI, which is responsible for the 4-thiouridine (S4) modification at the same position 8 in prokaryotic tRNAs.
Fig. 3.
Crystal structure of CDAT8. (A) The overall structure of the CDAT8 dimer (2.4 Å, Rwork/Rfree = 21.6%/26.2%) shows the head-to-head arrangement of the cytidine deaminase domains. Individual monomers are displayed in red and brown for clarity, and the active-site zinc is shown as yellow and brown spheres. (B) The active-site zinc is coordinated by His44, Cys67, Cys70, and a water molecule (blue). The catalytic glutamic acid (E46) is in close proximity to the active site. (C) Superimposition of APOBEC3G (PDB ID 3E1U, blue) onto the deaminase domain of CDAT8 (red). (D) Superimposition of the Bacillus anthracis ThiI (PDB ID 2C5S) ferredoxin-like and THUMP domains (blue) with the corresponding domains in CDAT8 (red) shows a high degree of structural homology. (E) The surface electrostatics of the CDAT8 monomer show a positively charged groove on the enzyme surface. A model of tRNA binding (black outline) shows position 8 of the tRNA adjacent to the enzyme active site.
CDAT8 forms a dimer with an extensive interaction interface of >1800 Å2 per monomer. The relative orientation between CDD and the FLD and THUMP domains is different in the two monomers (fig. S2). This asymmetric dimer appears to have a rigid, defined structure, as all four independent dimers in two different crystal forms are identical. The CDAT8 dimer interface is provided by both the CDD and FLD.
Dimerization of CDAT8 appears to facilitate the binding of tRNA and to orient it for deamination at position 8. We propose a model of tRNA interaction in which the tRNA acceptor stem binds to the dimer interface, with the 3′-CCA end at the THUMP domain of one monomer and U8 near the CDD active site of the other monomer (fig. S3). Because of the asymmetric dimer conformation, it appears that only one catalytic site is oriented appropriately for catalysis. Substantial evidence exists for this model of tRNA binding. First, the THUMP domain is known to bind tRNA (15, 16, 21) and the CDAT8 THUMP domain is structurally homologous to that of ThiI. Second, we determined that a tRNA substrate containing only the acceptor stem microhelix is sufficient for editing. Third, it has been shown that a shortened or blunt CCA end is deleterious to the activity of ThiI (22). Because 30 M. kandleri tRNAs are edited by a single deaminase, CDAT8 must recognize the conserved, sequence-independent features of tRNAs, such as the 3′-CCA overhang and the size and shape of the tRNA stems. Finally, a positively charged groove extends from the THUMP domain to the active site of the enzyme. The width of the groove formed between the two monomers could provide a snug fit for the acceptor stem; the length of the groove ensures that binding of the 3′-CCA at the THUMP domains places U8 adjacent to the active site of the CDD.
It is possible that C8 is the result of genetic drift. Alternatively, C8 may be beneficial for M. kandleri at the level of the tRNA gene, the primary tRNA transcripts, or the maturation steps that involve folding and modification of the tRNA. Because M. kandleri grows at extreme temperatures (up to 110°C), C8 may stabilize the genome at the tRNA gene or aid in tertiary folding and modification events of RNA molecules at such extreme conditions. Perhaps the 4-thiolation of U8 to S4U8 mediated by ThiI might be regulated by CDAT8, as C8 would not be a ThiI substrate. A C8 may also be beneficial at the level of DNA, as a T8C mutation might prevent the interaction of tRNA genes with mobile genetic elements [e.g., (23, 24)] at this otherwise fixed and convenient target.
Supplementary Material
Acknowledgments
We thank J. Yuan, P. O’Donoghue, and R. L. Sherrer for help and encouragement; K. O. Stetter for a gift of M. kandleri cells; and the staff at the Advanced Photon Source (beamline 24-ID), the National Synchrotron Light Source (beamlines X25 and X6A), and the Center for Structural Biology at Yale University. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (code 3G8Q). Supported by NIH grants GM22854 (D.S.) and AI078831 (Y.X.).
Footnotes
References and Notes
- 1.Teng B, Burant CF, Davidson NO. Science. 1993;260:1816. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 2.Gerber AP, Keller W. Science. 1999;286:1146. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- 3.Maas S, Gerber AP, Rich A. Proc Natl Acad Sci USA. 1999;96:8895. doi: 10.1073/pnas.96.16.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf J, Gerber AP, Keller W. EMBO J. 2002;21:3841. doi: 10.1093/emboj/cdf362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grosjean H, Constantinesco F, Foiret D, Benachenhou N. Nucleic Acids Res. 1995;23:4312. doi: 10.1093/nar/23.21.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinhauser S, Beckert S, Capesius I, Malek O, Knoop V. J Mol Evol. 1999;48:303. doi: 10.1007/pl00006473. [DOI] [PubMed] [Google Scholar]
- 7.Börner GV, Mörl M, Janke A, Pääbo S. EMBO J. 1996;15:5949. [PMC free article] [PubMed] [Google Scholar]
- 8.Rubio MA, et al. Proc Natl Acad Sci USA. 2007;104:7821. [Google Scholar]
- 9.Palmer JR, Baltrus T, Reeve JN, Daniels CJ. Biochim Biophys Acta. 1992;1132:315. doi: 10.1016/0167-4781(92)90168-y. [DOI] [PubMed] [Google Scholar]
- 10.Slesarev AI, et al. Proc Natl Acad Sci USA. 2002;99:4644. [Google Scholar]
- 11.Westhof E, Dumas P, Moras D. J Mol Biol. 1985;184:119. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- 12.Jones CN, Jones CI, Graham WD, Agris PF, Spremulli LL. J Biol Chem. 2008;283:34445. doi: 10.1074/jbc.M806992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sterner T, Jansen M, Hou YM. RNA. 1995;1:841. [PMC free article] [PubMed] [Google Scholar]
- 14.Smith AA, Carlow DC, Wolfenden R, Short SA. Biochemistry. 1994;33:6468. doi: 10.1021/bi00187a012. [DOI] [PubMed] [Google Scholar]
- 15.Aravind L, Koonin EV. Trends Biochem Sci. 2001;26:215. doi: 10.1016/s0968-0004(01)01826-6. [DOI] [PubMed] [Google Scholar]
- 16.Waterman DG, Ortiz-Lombardia M, Fogg MJ, Koonin EV, Antson AA. J Mol Biol. 2006;356:97. doi: 10.1016/j.jmb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 17.See supporting material on Science Online.
- 18.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. Nature. 2007;445:447. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 19.Chen KM, et al. Nature. 2008;452:116. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 20.Holden LG, et al. Nature. 2008;456:121. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. J Mol Biol. 2007;373:1243. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 22.Lauhon CT, Erwin WM, Ton GN. J Biol Chem. 2004;279:23022. doi: 10.1074/jbc.M401757200. [DOI] [PubMed] [Google Scholar]
- 23.Marschalek R, Brechner T, Amon-Bohm E, Dingermann T. Science. 1989;244:1493. doi: 10.1126/science.2567533. [DOI] [PubMed] [Google Scholar]
- 24.She Q, Brugger K, Chen L. Res Microbiol. 2002;153:325. doi: 10.1016/s0923-2508(02)01331-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.