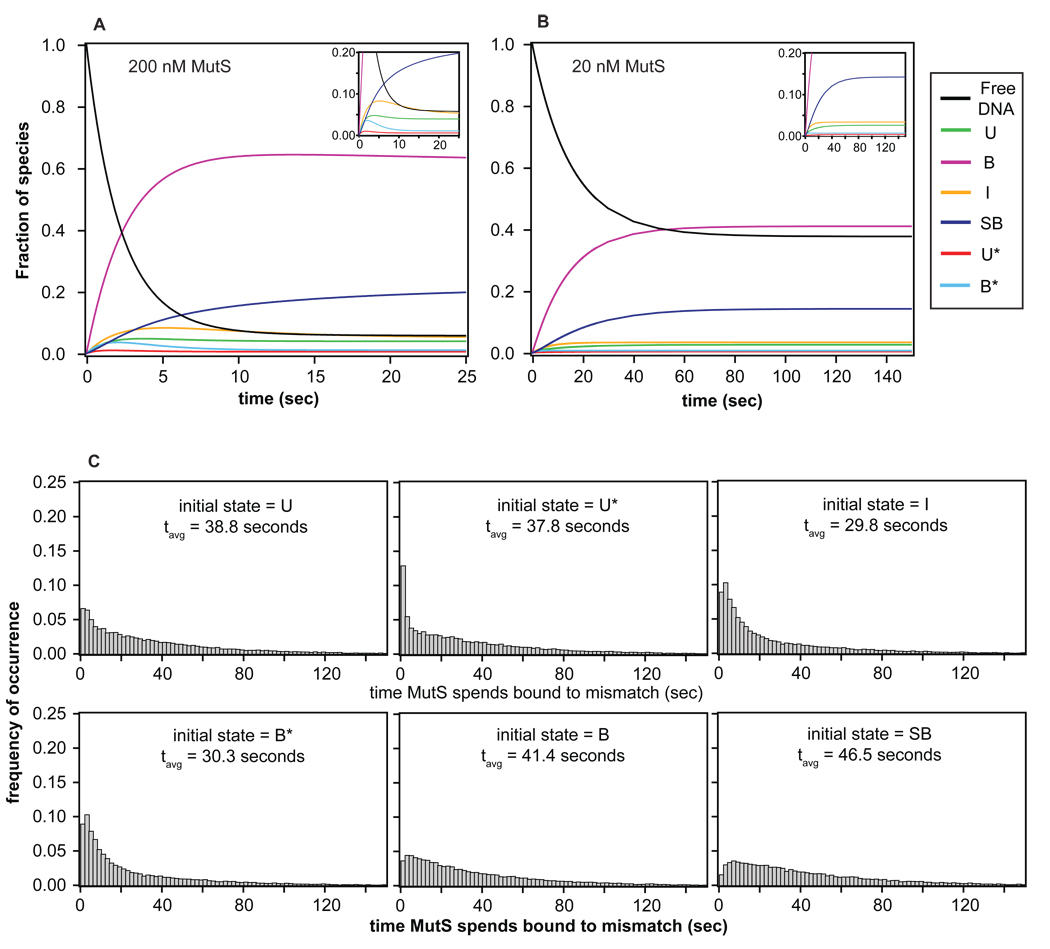
Figure 8.
Simulations of bulk and single molecule kinetic behavior. (A–B) Evolution of each species as a function of time for an ensemble of molecules using the similarity transform procedure. The inset shows the evolution of states with fractions less than 0.20. Results, shown as the fraction of species, reveal that the system reaches equilibrium in about 20 seconds at 200 nM MutS and in about 120 seconds at 20 nM MutS. The fraction of each species observed at equilibrium in the simulations are consistent with those determined from the experimental rates (Figure 7), confirming the accuracy in the FRET TACKLE kinetic calculations. (C) Distributions of the time MutS spends bound to the DNA generated from Monte Carlo simulations of single binding event (MCSBE) when each of the 6 different states is used as the initial state of the simulation. These results directly show that the time MutS spends bound to the DNA is highly dependent on the state in which the complexes start, where occupancy is highest when the starting state is the stable bent conformation B, and occupancy is the lowest when the starting state is the intermediately bent state I.