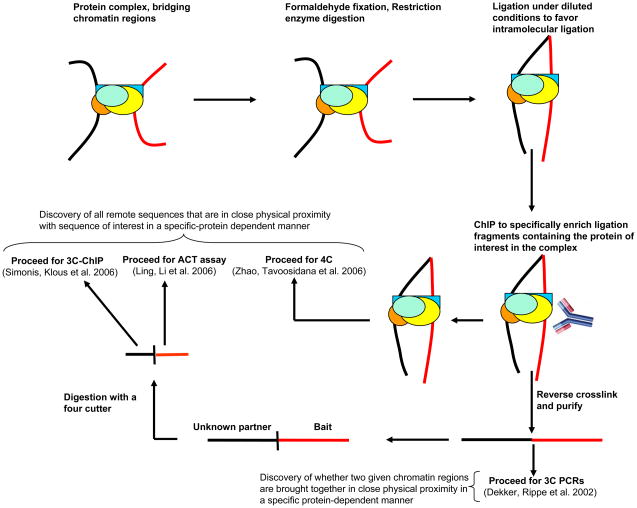
Figure 2.
Further applications and the future of Combined 3C-ChIP-Cloning (6C) methodology. 6C method can be combined with other techniques to identify the entire “interactome” in the nucleus for any gene or chromatin region of choice that is mediated by a specific protein of interest. Following the ChIP, the samples could be processed for 4C analysis (Zhao, Tavoosidana, Sjolinder, Gondor, Mariano, Wang, Kanduri, Lezcano, Sandhu, Singh et al. 2006) or reverse crosslinked, purified, digested with a four cutter of choice and further subjected to either 3C-chip (Simonis, Klous, Splinter, Moshkin, Willemsen, de Wit, van Steensel and de Laat 2006) or ACT assay (Ling, Li, Hu, Vu, Chen, Qiu, Cherry and Hoffman 2006). This technique could also be used to investigate whether two known chromatin regions are brought in close physical proximity by a protein of interest. For this purpose, subsequent to the reversal of crosslinking and purification, the amplification criteria used in the original 3C assay should be followed (Dekker, Rippe, Dekker and Kleckner 2002).