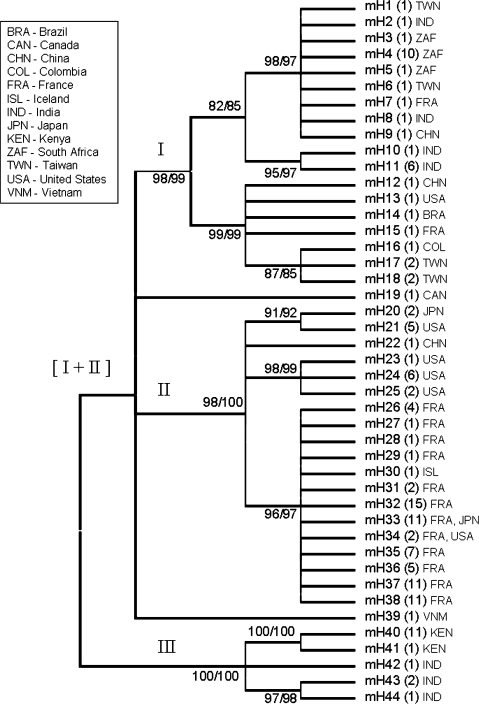
FIG. 3.
Phylogenetic analysis of Caenorhabditis briggsae mitochondrial haplotypes. Unrooted 80% bootstrap consensus cladogram was constructed using 1,694 bp of mtDNA, including both Ψnad5-1 and Ψnad5-2, analyzed in all 141 isolates. The 44 unique mtDNA haplotypes were used for phylogenetic analysis. Ψnad5-2 is absent in Clade III isolates—the gapped region in Clade III isolate sequences was treated as a single event in phylogenetic analyses (complete deletion parameters in MEGA4). Bootstrap values for maximum parsimony (left) and NJ (right) methods (1,000 replicates performed for each) are indicated to the left of the corresponding node. Numbers in parentheses next to haplotype designators show the numbers of isolates with that haplotype. Three-letter country code designators (United Nations designators) are also provided for general geographic information—see supplementary table 1 (Supplementary Material online) for more detailed information on haplotypes and geographic locations, especially for isolates collected from islands. Complete phylograms from NJ and maximum parsimony analyses are shown in supplementary figs. 1 and 2 (Supplementary Material online), respectively.