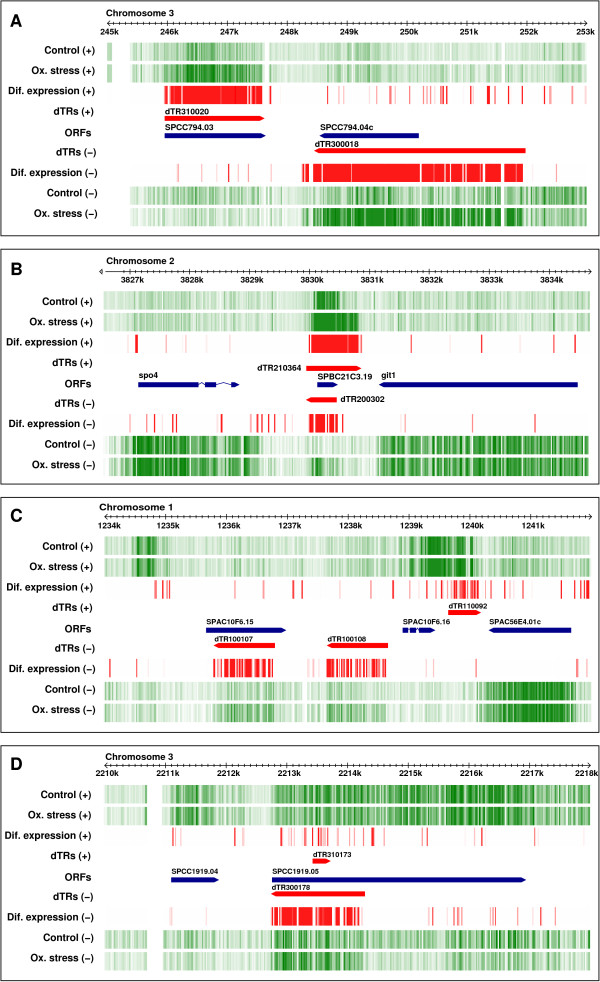
Figure 5.
Browser visualization of differential expression. Vertical green lines represent transcription from both DNA strands (indicated by + or -) in control S. pombe cells and in cells under oxidative stress. Darker green indicates a higher level of expression. Red vertical lines indicate a differential over- or under-expression level between both physiological conditions greater than 1.75-fold. Differences below this level are not shown. ORFs are indicated by blue bars pointing towards the direction of transcription. Differentially transcribed regions (dTRs) showing differential expression are represented by red bars. A) Overexpression of dTR310020 and dTR300018 from complementary strands encompassing the SPCC794.03 and SPCC794.04c genes under oxidative stress. B) Overexpression of dTR210364 and antisense dTR200302 from complementary strands encompassing the SPBC21C3.19 gene. C) Overexpression of non-annotated dTR100108 and dTR110092. Antisense transcription is also detected across SPAC10F6.15 under conditions of oxidative stress. D) Strong overexpression of antisense RNA (dTR300178) from a 1.5 kb region at the 5' end of the SPCC1919.05 gene. Quantitative data for all differentially expressed annotated and non-annotated dTRs across the complete genome are shown in Additional File 2. The data can be accessed from the genome browser on our website http://genomics.usal.es/TMADE/browser.