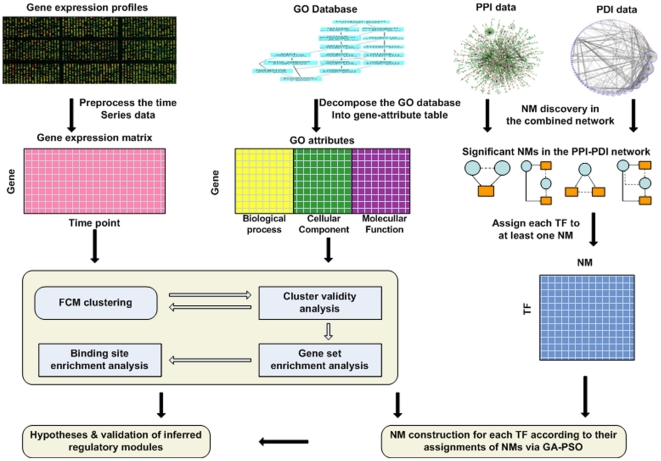
Figure 1. Schematic overview of the computational framework used for the network motif regulatory module inference.
Gene expression patterns were first clustered into biologically meaningful groups by FCM; GO category information of genes was used to determine the optimal cluster number. To evaluate the gene clusters, GSEA was performed on the optimal clusters. Additionally, significant network motifs detected in the combined network of PPI and PDI were then assigned to each transcription factor. After the gene clusters are formed and transcription factors were assigned to network motif categories, the connections between transcription factors and gene clusters were inferred by training RNNs that mimic the topology of the network motifs that transcription factors are assigned to. Finally, the inferred network motifs were validated by BSEA and literature results.