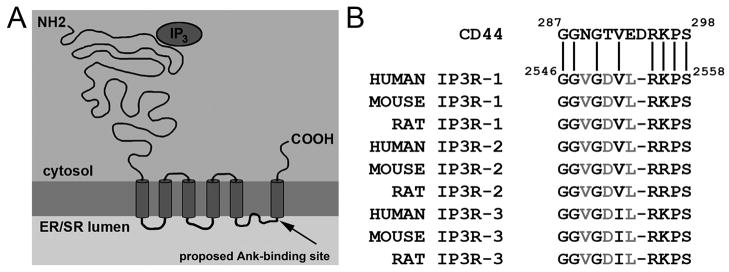
Figure 1. Domain organization of InsP3 receptor and InsP3 receptor intermolecular interactions.
A, Domain organization of the InsP3 receptor monomer, depicting membrane topology and cytoplasmic and lumenal protein orientation. Inositol 1,4,5-trisphosphate (InsP3; oval) activates Ca2+ release via binding to an N-terminal region. Arrow denotes location of previously proposed ankyrin-binding motif in C-terminal domain of InsP3 receptor (see Figure 1B). Note that these residues (rat 2546–2558) are located in the ER/SR lumen, inconsistent with an interaction with cytosolic ankyrin. B, Ankyrin-binding site in InsP3 receptor identified by sequence similarity with CD44 ankyrin-binding motif. Minimal ankyrin-binding residues on CD44 [Bourguignon and Jin, 1995]. Amino acid sequences below CD44 denote sequence homology of residues in InsP3 receptor C-terminus to the CD44 sequence.