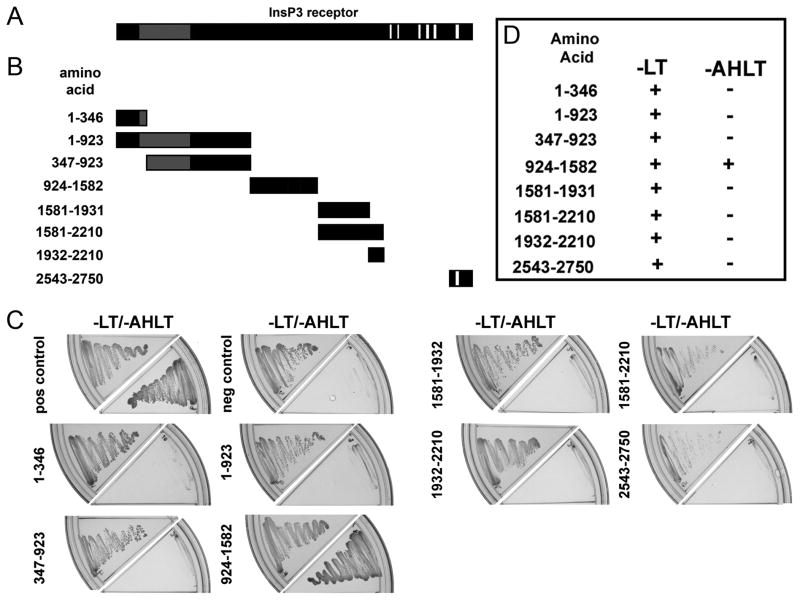
Figure 2. Identification of requirements on InsP3 receptor for ankyrin-B association.
A, Diagram of the InsP3 receptor monomer [da Fonseca et al., 2003]. Gray box represents InsP3 binding domain and white boxes represent transmembrane domains. B, InsP3 receptor yeast two-hybrid constructs. Fragments of the cytoplasmic regions of the InsP3 receptor monomer used in the initial assessment of the structural requirements of InsP3 receptor for interaction with ankyrin-B. InsP3 receptor construct 2543–2750 contains the previously predicted ankyrin-binding domain [Bourguignon and Jin, 1995]. Constructs were designed based on identified structural folding boundaries defined by limited trypsin digestion [Yoshikawa et al., 1999]. C, AH109 yeast were co-transformed with ankyrin-B membrane-binding domain (bait) and a unique InsP3 receptor construct (prey). Co-expression of bait and prey proteins was confirmed by inoculating onto -LT media. Positive interactions were determined by growth on -AHLT media after five days incubation. TD1-1 served as bait for both the positive and negative controls, while pVA3 and pLAM5 were used as prey for the positive and negative controls, respectively. D, Summary data for ankyrin-B membrane-binding domain-InsP3 receptor interactions.