Fig. 4.
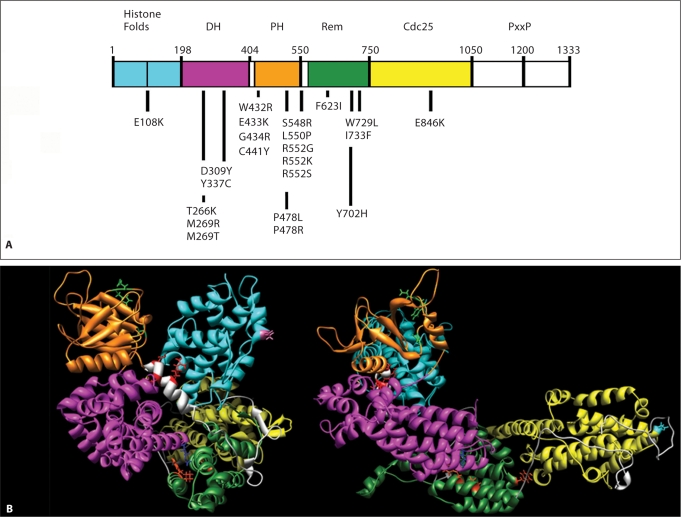
SOS1 domain structure and location of affected residues in NS. A SOS1 missense mutations are positioned below the cartoon of the SOS1 protein with its functional domains indicated above. Abbreviations: DH, Dbl homology domain; PH, plekstrin homology domain; Rem, RAS exchanger motif; PxxP, proline-rich motif. B Location of the mutated residues on the 3-dimensional structure of SOS1. The functional domains are color-coded as follows: cyan, histone folds; magenta, DH; orange, PH; green, Rem; yellow, Cdc25. Residues affected by mutations are indicated with their lateral chains (violet, histone folds; blue, DH; green, PH; red, helical linker; orange, Rem; cyan, Cdc25). Based on Sondermann et al. [2005], who utilized structural data and computational modeling.