Abstract
Many MHC class I binding peptides are generated as N-extended precursors during protein degradation by the proteasome. These peptides can be subsequently trimmed by aminopeptidases in the cytosol and/or the ER to produce mature epitope. However, the contribution and specificity of each of these subcellular compartments in removing N-terminal amino acids for antigen presentation is not well defined. Here we investigate this issue for antigenic precursors that are expressed in the cytosol. By systematically varying the N-terminal flanking sequences of peptides we show that the amino acids upstream of an epitope precursor are a major determinant of the amount of antigen presentation. In many cases MHC class I binding peptides are produced through sequential trimming in both the cytosol and ER. Trimming of flanking residues in the cytosol contributes most to sequences that are poorly trimmed in the ER. Since N-terminal trimming has different specificity in the cytosol and ER, the cleavage of peptides in both of these compartments serves to broaden the repertoire of sequences that are presented.
Keywords: Antigen Presentation/Processing, Antigens/Peptides/Epitopes, MHC
Introduction
Major histocompatibility complex (MHC) class I molecules present to the immune system a large variety of peptides generated by intracellular protein degradation. This allows identification and elimination of cells synthesizing abnormal or “foreign” proteins, which may be a result of mutation or infection by virus (1–5).
Peptides that can stably bind MHC-I molecules are generally 8, 9, or 10 residues in length, and are generated by a complex pathway involving many components in several cellular compartments. The first step in this pathway is degradation of proteins by the proteasome in the nucleus or cytosol. This multi-subunit protease is capable of generating mature epitopes (roughly <5% of product (6)). However, the majority of epitope-containing peptides (15 to 25 % of proteasome product (6)) are extended on the N-terminus and are incapable of MHC binding unless processed further (7). Within the cytosol there are a large number of aminopeptidases and endopeptidases that have the potential to create or destroy MHC binding peptides from the precursors generated by the proteasome (8–17). Within seconds the majority of peptides in the cytosol are hydrolyzed into amino acids which can then be reused in synthesis of new proteins (18). However some escape destruction and are shuttled by the transporter associated with antigen presentation (TAP) into the ER. In addition, some MHC class I-presented peptides may be generated through the trimming of long antigenic precursors in the cytosol (19). TAP translocates peptides longer than 16 residues inefficiently in vitro (20), therefore trimming of long precursors by peptidases within the cytosol may also be critical for generation of peptides suitable for TAP binding (21).
A substantial amount of the trimming of N-extended precursors is performed in the ER by the IFN-γ inducible metallopeptidase ER aminopeptidase 1 (ERAP1) (ER aminopeptidase associated with antigen processing; ERAAP) (19, 22, 23). ERAP1 prefers substrates 9–16 residues long (19, 24), which corresponds to the length of peptides efficiently transported by TAP (20). ERAP1 appears to be particularly adapted to generate peptides for MHC class I presentation because it rapidly trims precursors down to 8 or 9 residues before cleavage slows or stops completely. Thus, unlike other aminopeptidases that will continually cleave peptides until only one free amino acid remains, ERAP1 acts with a molecular ruler to generate peptides of the optimal length for MHC class I binding (24). It has also been suggested that ERAP1 cooperates with MHC class I molecules to generate mature epitopes although the available data do not distinguish whether MHC class I molecules are actually involved in the trimming process or only bind and protect mature epitopes from destruction (25). In cells lacking ERAP1, many antigenic precursors are trimmed poorly if at all in the ER (19, 22, 26–28), and mice lacking ERAP1 had markedly different presentation of a variety of epitopes (26–29), leading to differences in immunodominance hierarchies (26, 29, 30). However, precursor peptides generated in the cytosol are still presented to various extents in ERAP1-deficient cells, indicating that some trimming of presented peptides occurs in the cytosol (19).
Schatz et al have recently investigated the role that the ER and cytosol may play in epitope generation in cell extracts (31), and we have examined the role of ER trimming in intact cells (32). These studies suggest that trimming in the ER plays an important part in generating antigenic peptides, but does not account for all peptide processing. Understanding the contribution and specificity of non-proteasomal proteases in vivo in the generation of CTL epitopes should enhance our understanding of epitope generation and modeling of this process. This led us to initiate the present study to systematically examine the specificity of trimming N-terminal sequences from antigenic precursors in vivo.
Material and Methods
Plasmids
To express N-terminal S-L (OVA254–264) precursors in the cytoplasm, synthetic complementary oligonucleotide primers (Integrated DNA Technologies, Coralville, IA) were annealed and inserted into pUG-1 (11). This generates a plasmid consisting of Ub with (X)n-S-L (where n is a variable number of residues) fused to the C-terminus. The oligonucleotides for cytoplasmic expression were, for Ub-XXS-L, 5′-XXX XXX TCC ATC ATC AAC TTC GAG AAG CTG TAA TAGCTGCAGGATCC -3′; and for Ub-4N-X-S-L (Ub-LEQLXS-L), 5′-CTG GAG CAG CTG XXX TCC ATC ATC AAC TTC GAG AAG CTG TAA TAGCTGCAGGATCC-3′. A Pst I site (CTGCAG) was used for selection. XXX is a codon that encodes for an amino acid. The most common codons for each amino acid, based on frequency of usage of each codon (per thousand) in human coding regions, were used; ala (A), GCT; arg (R), CGC; asn (N), AAC; asp (D), GAC; cys (C), TGT; gln (Q), CAG; glu (E), GAG; gly (G), GGC; his (H), CAC; ile (I), ATC; leu (L), CTG; lys (K), AAG; met (M), ATG; phe (F), TTC; ser (S), TCC; thr (T), ACC. All plasmids were sequenced to confirm correct sequences and reading frames. Generation of ss XXS-L plasmids have been described previously (32).
Cells
HeLa-Kb and HeLa-Kb-ICP47 (HeLa-Kb-A3-47 cells (HeLa cells stably transfected with H-2Kb, HLA-A3 and ICP47)) cells have been described previously (19, 33). Mouse embryonic fibroblasts (MEFs) were prepared from wild-type C57BL/6 (two independent lines) and ERAP1 knockout mice (two independent lines) as previously described (26).
siRNA
Cells were transfected with siRNA for both ERAP1 and control mTOP (directed against murine TOP in a region that differs from human TOP) using oligofectamine (Invitrogen, CA) as previously described (19).
Antibodies and flow cytometry
HeLa cell lines were transiently transfected with plasmid (0.25–1 μg) using TransIT HeLa Monster (Mirus, Madison, WI) according to the manufacturer’s protocol. MEFs were transfected with FuGene6 (Roche) according to the manufacturer’s protocols. After transfection, cells were incubated for 24–48hrs and H-2Kb-S-L complexes on the cell surface were detected using mAb 25.D1.16 (34). The cells were analyzed by flow cytometry on a FACSCalibur apparatus (Becton Dickinson, San Jose, CA) with CellQuest (Becton Dickinson) or FlowJo (Tree Star, San Carlos, CA) software, gating on GFP-expressing cells to limit the analysis to transfected cells (Supplementary Figure 1). Unless otherwise stated, data are representative of three independent experiments.
Results
The effect of N-terminal flanking residues on the presentation of cytosolic precursors
Since antigenic epitope precursors are first generated in the cytosol which contains resident aminopeptidases that can trim peptides, we sought to analyze the specificity of trimming of N-extended precursors in this compartment.
In order to systematically examine the specificity of trimming of N-extended precursors of MHC class I-presented peptides we generated a series of minigene constructs containing the model epitope SIINFEKL (S-L) (the immunodominant H-2Kb-restricted epitope from chicken ovalbumin). Minigenes were constructed to encode 19 different amino acids (proline was excluded for reasons described below) on the N-terminus of the mature epitope. In order to amplify any differences between constructs in the rate of removal of their flanking residues, they were expressed with two identical amino acids upstream of the epitope, generating XXS-L, where “X” represents any amino acid. These sequences were ligated in frame to the 3′ region of ubiquitin (Ub) so that they would be expressed as fusion proteins (Ub XXS-L) in the cytosol. Ubiquitin C-terminal hydrolases in cells efficiently and precisely cleave the peptide bond between the ubiquitin moiety and any fused sequence (unless the residue flanking the scissile bond is a proline (35)), which is why proline was omitted from our series); thus when expressed in cells these constructs efficiently generate XXS-L in the cytoplasm and avoids the need for an initiating methionine on the N-terminus of the antigenic peptide. The Ub XXS-L sequences were cloned into a plasmid that bicistronically expressed GFP, allowing the direct correlation of GFP and Ub XXS-L expression.
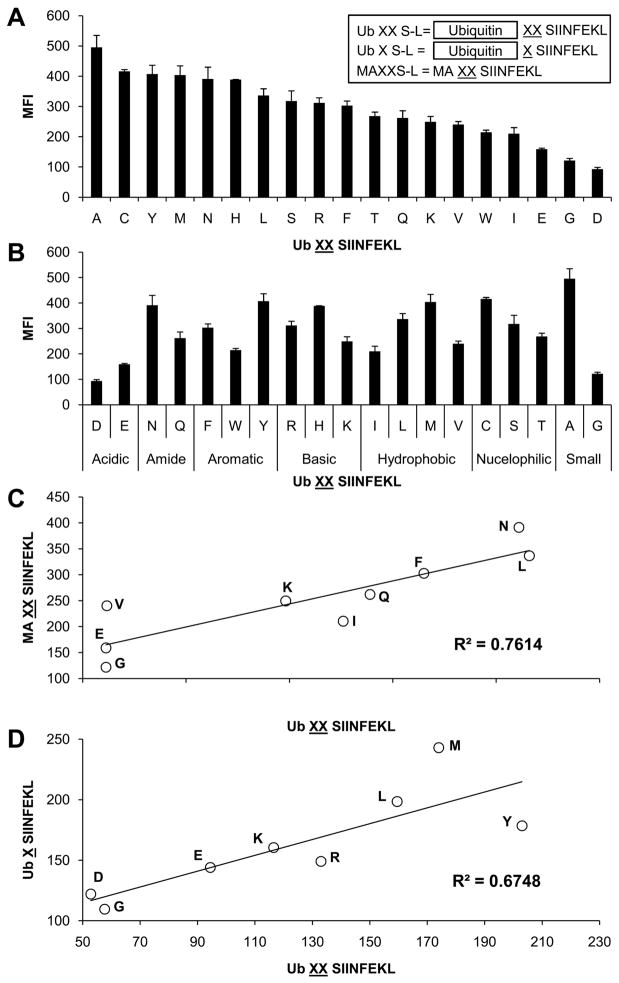
The Ub XXS-L constructs were transiently transfected into HeLa-Kb cells and the amount of S-L-Kb complexes on the cell surface of cells expressing similar levels of GFP was quantified by staining with the monoclonal antibody 25.D1.16, which recognizes the combination of H-2Kb and S-L (34). When N-extended precursor peptides are expressed in cells, only the mature epitopes are presented on H-2Kb and detected by 25.D1.16 (19, 21). This assay gave highly reproducible results between replicate groups and independent experiments. This analysis revealed that N-terminal flanking amino acids had a marked influence on the levels of presented peptide detected on the cell surface (Figure 1A). S-L from all constructs was presented, but at different levels, such that there was a highly reproducible hierarchy of presentation that depended on the specific N-terminal residues. Amino acids such as alanine, cysteine, tyrosine and methionine appear to be efficiently removed from precursors to generate the mature S-L epitope. In fact 16 of the 19 amino acids (including alanine) were associated with presentation that was ≥ 40% of that observed with Ub AAS-L (the construct leading to the greatest presentation), demonstrating that many amino acids are removed rather efficiently. Only glutamate, aspartate and glycine were associated with lower presentation, <40% of that observed with Ub AAS-L. However, although the acidic amino acids glutamate and aspartate appear to be associated with low presentation, the hierarchy of presentation did not otherwise correlate with the chemical nature of the side chain. Thus among the various classes of amino acids (basic, hydrophobic, etc.) there was a broad range of efficiencies of presentation within each group (Figure 1B).
Figure 1. Antigen presentation in HeLa-Kb cells transfected with cytoplasmic targeted N-terminal S-L precursors.
Cells were transfected with Ub XS-L, Ub XXS-L or MAXXS-L (A inset), where X represents one of the twenty amino acids. H-2Kb-S-L presentation was determined by staining cells with 25.D1.16. Data arranged in order of efficiency of presentation, from highest to lowest, left to right (A) or grouped into chemical nature of the amino acid side chain (B). Graphs represent the average MFI of two independent transfections. Error bars represent the difference within each group. Correlation of S-L presentation from Ub XXS-L and MAXXS-L (C) or Ub XS-L (D).
If the ubiquitin C-terminal hydrolases were to cleave the various Ub XXS-L constructs imprecisely or at different rates, this could contribute to the differential presentation of the various Ub XXS-L antigens. Although these proteases are thought to cleave precisely and independently of the P′ residue (35) we sought to evaluate this issue. To attempt to exclude the possibility that differences in presentation between the various constructs was due to the specificity of the ubiquitin hydrolases, we sought to generate XXS-L in another way. For these experiments we generated a subset of minigenes that consisted of a Kozac consensus sequence followed by an initiating methionine and the sequence AXXS-L (we excluded X=M from this analysis because of the potential for multiple translation initiation sites) were X encoded amino acids which were processed efficiently (L and N) and inefficiently in the cytoplasm (E, G and I) as well as amino acids which lie in between these two extremes (F, K, Q and V). When these constructs were transfected into HeLa-Kb cells, they are trimmed and presented on H-2Kb (Figure 1C). Again there was a hierarchy of presentation of the various constructs and importantly this hierarchy was essentially the same as that observed with Ub XXS-L (Figure 1C). The concordance of the data from the Ub XXS-L and MAXXS-L constructs argues strongly that the hierarchy in presentation is not due to differences in the generation of precursors, but rather is due to processing of the precursors to the mature peptide.
Given that the identity of the amino acid repeated twice N-terminally of an epitope markedly influences its presentation when expressed in the cytosol, we next examined whether a single amino acid showed the same effect as the corresponding doublet. To this end we generated a subset of Ub XS-L constructs which encoded amino acids which were processed efficiently (L, M and Y) and inefficiently in the cytosol (D, E and G) as well as amino acids which lie in between these two extremes (K and R). The data obtained with these constructs (Figure 1D) are largely consistent with those obtained with the corresponding doublet although the absolute differences in the level of presentation were smaller presumably due to the reduction in the number of residues that needed to be removed to generate mature epitope (Figure 1A).
Ub XXS-L trimming by ERAP1
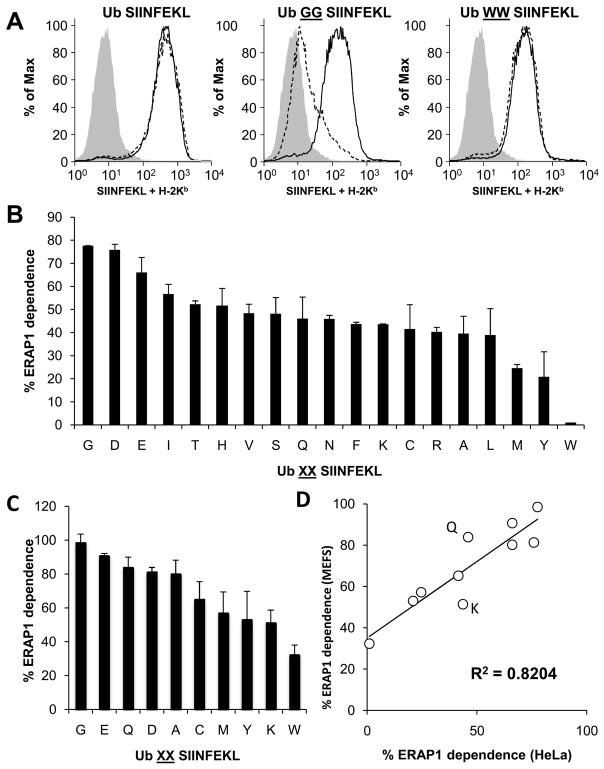
Antigenic precursors produced in the cytosol could potentially be trimmed in the cytosol and/or in the ER after transport by TAP. In the ER, the aminopeptidase ERAP1 appears to be the only important peptide-trimming enzymatic activity in the ER of HeLa cells as demonstrated using S-L precursors with the natural flanking sequence (LEQLE) (19, 22). We therefore sought to determine how much ERAP1 contributes to the presentation of the various Ub XXS-L constructs. These precursors were transfected into HeLa-Kb cells treated with ERAP1-specific siRNA under conditions where ERAP1 protein expression is reduced by at least 90% (19) and presentation was compared to cells treated with a control irrelevant siRNA.
Ubiquitin C-terminal hydrolase cleavage of the control Ub S-L in the cytosol generates mature S-L directly. As this mature epitope requires no ER trimming for presentation its presentation should not be dependent on ERAP1. Treatment with ERAP1 siRNA did not affect presentation from this precursor (Figure 2A), demonstrating that the siRNA does not cause off-target effects on antigen presentation. In contrast, the presentation of S-L from most of the Ub XXS-L constructs was reduced in ERAP1-silenced cells, suggesting a role for this enzyme in epitope generation from cytosolic precursors. Interestingly, there was a range of ERAP1-dependent presentation from these various N-terminally extended precursors. For example, presentation from Ub GGS-L was severely reduced in ERAP1-silenced cells, whereas presentation from Ub WWS-L was unaffected, (again a finding suggesting no off-target effects of the siRNA-treatment) (Figure 2A). Dependence on ERAP1 was approximately similar for the other 17 constructs and lay between these two extremes (Figure 2B). Overall, the magnitude of the ERAP1 dependence ranged from 0 to 80%.
Figure 2. Effect of ERAP1 knockdown on S-L presentation from cytosol targeted XXS-L.
HeLa-Kb cells were treated with siRNA targeting ERAP1 or control (ctrl) siRNA then transfected with Ub XXS-L. On day three S-L presentation was determined. FACS showing cells stained with 25.D1.16 following transfection with Ub S-L, Ub GGS-L and Ub WWS-L; cells treated with ctl siRNA (Solid line) or ERAP1 siRNA (dashed line). Shaded histogram, background. Percentage of S-L presentation that is ERAP1 dependent, following HeLa transfected with Ub XXS-L (B). Arrange from highest to lowest, left to right. Calculated from the MFI of cells treated with ctrl (100%) or ERAP1 siRNA. Graphs represent the average percentage of two independent experiments. (C) ERAP1 dependence of Ub XXS-L contructs transfected into MEFs (ERAP1 −/− MEFS compared with ERAP+/+ MEFS) arranged highest to lowest (graphs represent the average percentage of three independent experiments). (D) Scatter graph showing correlation of ERAP1 dependence between MEFs and HeLa cells.
The presentation that occurs in ERAP1-silenced cells presumably reflects trimming occurring in the cytosol and possibly some contribution from residual ERAP1. The large difference in ERAP1 dependence of the various XXS-L constructs suggests that there is sequence specificity for trimming by other peptidases responsible for the generation of S-L. Although ERAP1 siRNA reduces ERAP1 expression by at least 90% (19), a small amount of residual ERAP1 could still process precursors under these conditions. To test the potential contribution of residual ERAP1, we measured presentation of a subset of peptides in mouse embryonic fibroblasts (MEFs) prepared from wild-type (ERAP1 +) or ERAP1 knockout mice, in which ERAP1 is completely absent. Again, the dependence on ERAP1 for presentation was different for different constructs (Figure 2C), and their pattern of dependence on ERAP1 correlated strongly between MEFs and HeLa cells (R2 = 0.82) (Figure 2D). The magnitude of ERAP1 dependence in MEFs was somewhat greater than in HeLa cells, ranging from about 30% to nearly 100%, suggesting that the small amounts of ERAP1 in siRNA-treated Hela cells may contribute slightly to peptide processing. Alternatively, mouse embryonic fibroblasts may have a slightly different spectrum of peptidases such that ERAP1 is more important than in Hela cells. In any case, the strong correlation between HeLa cells and MEFs suggests that peptide processing is remarkably similar not only between different cell types, but between different species.
Interestingly, even in MEFs, the absence of ERAP1 only modestly reduced the presentation of the Ub WWS-L construct. Therefore, a cytosolic peptidase can play a dominant role in removal of tryptophan residues upstream of S-L prior to transport into the ER.
Presentation and ERAP1-dependence of precursors expressed in the cytosol vs. the ER
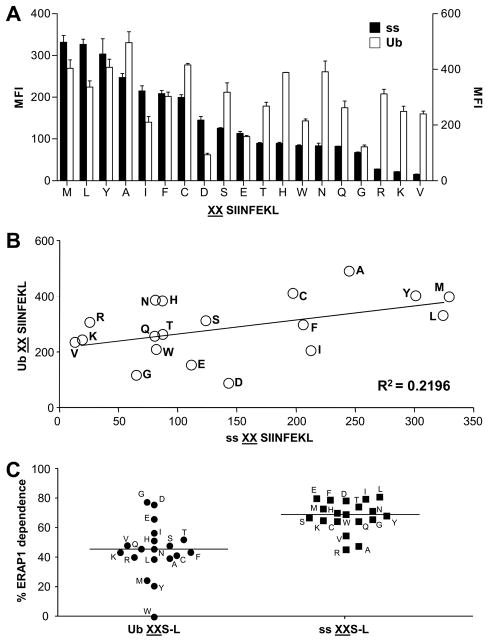
It was of interest to compare the presentation of XXS-L constructs in the cytosol versus ER. To do this we compared the presentation of the Ub XXS-L peptides, to ER-targeted peptides that we had previously generated and characterized in HeLa Kb cells stably transfected with ICP-47 (32). ICP-47 blocks the TAP transporter and prevents cytosolic peptides from gaining access into the ER (36), allowing specific analysis of ER specific trimming. These ER-targeted constructs consisted of the XXS-L sequences that were fused to an N-terminal signal sequence (ss XXS-L). Upon synthesis these constructs are transported via the sec61 translocon into the ER where the leader peptide is removed by the signal sequence peptidase; our previous studies found that differences in the presentation of these constructs were not due to differences in the efficiency of cleavage of the various constructs by the signal sequence peptidase (32).
We had previously examined the presentation of the ER-targeted constructs in HeLa Kb cells in which TAP was inhibited (32). In HeLa Kb cells with a functional TAP transporter the difference between highest and lowest presentation of Ub XXS8L and ss XXS8L constructs was much greater for the ER-targeted precursors than the cytosolic precursors, and presentation from many of the epitope precursors was greater in relative terms if expressed in the cytosol than if targeted to the ER (Figure 3A). As a result when the averages of the results obtained in Figure 3A are plotted there is a poor correlation between presentation of the same constructs targeted into ER compared to the cytosol (Figure 3B). Presentation from cytosol precursors is probably the result of a combination of editing in the cytosol and the ER (Figure 2), whereas processing of ER precursors is only dependent on the specificities of the aminopeptidase(s) in this compartment (32). ERAP1 appears to play a major role in ER precursor editing in HeLa cells stably transfected with ICP-47 (32) as well as in many cases of cytosolic precursors in HeLa cells with an active TAP transporter (Figure 2). We next compared the role of ERAP1 in processing of cytosolic precursors (Figure 2) to that of ER precursors in HeLa cells with an active TAP transporter. Figure 3C shows that all of the ER targeted precursors (including WWS-L) rely much more heavily on the activity of ERAP1 than the majority of the cytosolic precursors. Thus, a large percentage of presentation from cytosolic precursors is due to an ERAP1-independent component, and the differences in presentation efficiency must reflect trimming within the cytosol.
Figure 3. Comparison of precursors expressed in the cytosol and the ER.
HeLa-Kb cells were transfected with ER (ss; black bars) or cytosol (Ub; white bars) targeted XXS-L and stained with 25.D1.16 (A). Data arranged in order of efficiency of presentation from ER targeted precursors, from highest to lowest, left to right. (B) Correlation of S-L presentation from Ub XXS-L and ss XXS-L (from the mean data in (A)). Following treatment with ctrl or ERAP1 siRNA, HeLa-Kb cells were transfected with ss XXS-L or Ub XXS-L. (C) The percentage of S-L presentation that is ERAP1 dependent was calculated (100-((MFI with ERAP1 siRNA/MFI with ctrl siRNA)*100). The Ub XXS8L data in C come from Fig 2B and are re-graphed for comparison with the new ss XXS8L data. Data represent the average percentage of two independent experiments
Presentation from precursors with and without TAP activity
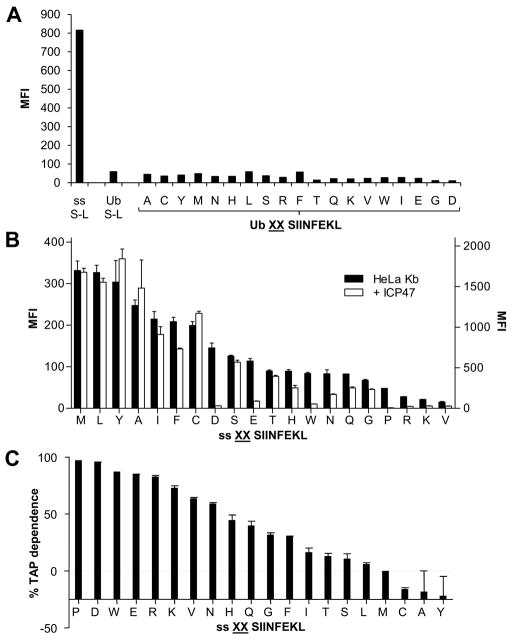
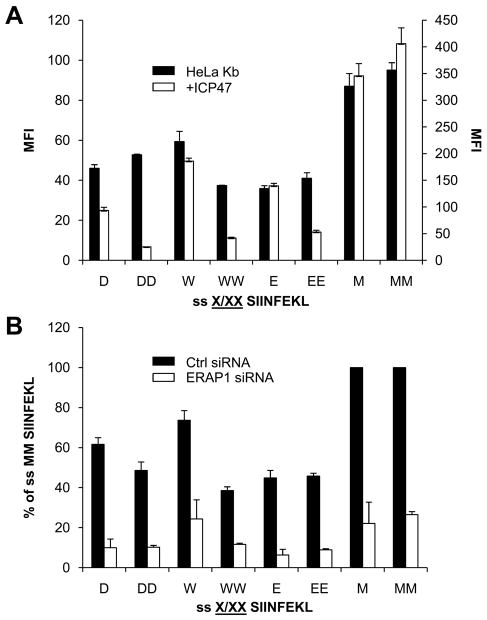
To further assess the compartment in which the various cytosolic or ER precursors are trimmed, we compared presentation of S-L in HeLa Kb or HeLa-Kb-ICP47 (HeLa cells stably transfected with ICP47 (33)). When SIINFEKL, the mature epitope was expressed within the cytoplasm (Ub S-L) no presentation was observed, indicating efficient TAP inhibition, while ER-targeted S-L (ss S-L), which bypasses the need for TAP transport, was presented efficiently (Figure 4A). As expected, the presentation of all the cytosolic precursors (Ub XXS-L constructs) was essentially completely inhibited in HeLa-Kb-ICP47 cells, since these cytosolic peptides require TAP for transport into the ER. On the other hand, presentation of S-L from most ER-targeted precursors was efficient (Figure 4B). Unexpectedly, however, presentation of S-L from many ER-targeted constructs (for example those with D, W and E) was partially to almost completely inhibited by ICP47 (Figures 4B and 4C, implying that presentation of these ER-targeted precursors was still at least partially TAP-dependent.
Figure 4. Affect of TAP inhibition on S-L presentation from cytosol and ER precursors.
HeLa-Kb-ICP47 cells were transfected with Ub XXS-L for 24hrs and stained for surface H-2Kb-S-L (A). ER targeted S-L (ss S-L) and cytoplasmic S-L (Ub S-L) were used as a positive control and negative control respectively. (B) Comparison of S-L presentation from ER targeted precursors following transfection of HeLa-Kb cells (black bars) and HeLa-Kb-ICP47 cells (white bars). Graph represents the average percentage presentation of two independent wells using the MFI of cells transfected with ss MMS-L as 100% presentation. Error bars represent the difference within each group. (C) Using the data in (B) the percentage TAP dependence of each ss XXS-L construct was calculated.
X/XXS-L cytosol dependence
Several studies have raised the possibility of retrotranslocation of epitope precursors from the ER into the cytoplasm for trimming followed by re-import of peptides into the ER for presentation (37–39). In this study some of the ER-targeted precursors (ss XXS-L) that show a strong reliance on the cytosol for presentation (i.e. are inhibited by ICP47, Figure 4B), also require ERAP1 for processing (Figure 3C). This suggests that epitope production may be dependent on trimming in both the cytosol and ER. Assuming that the ER targeted construct is not mistargeted into the cytosol then the epitope precursor must be retrotranslocated into the cytoplasm for processing. Our findings thus far suggest that for ER-targeted precursors that require ERAP1 and TAP either these XXS-Ls are poor substrates for ERAP1 and must be converted into XS-L in the cytosol (XXS-L → XS-L) for ERAP1 dependent removal of X (XS-L→S-L), or that following ERAP1-dependent generation of XS-L in the ER (XXS-L → XS-L), S-L must be generated in the cytosol (XS-L→S-L) because these XS-Ls are poor substrates for ERAP1. In order to distinguish between these possibilities we compared presentation from ss D/W/ES-L and ss DD/WW/EES-L (the latter of which is dependent on both ERAP1 and the cytosol for presentation (Figure 3C, 4B, 4C)) in HeLa-Kb cells with and without ICP47. ss MS-L and ss MMS-L (the latter of which is ERAP1-dependent and cytosol-independent (Figure 3C, 4B, 4C)) were used as controls. Presentation from both ss MS-L and ss MMS-L was not TAP dependent, implying that both M and MM can be removed efficiently in the ER for S-L production (Figure 5A). Interestingly, although presentation from ss DD/WW/EES-L was inhibited when TAP was inactivated, as shown previously, presentation from ss W/ES-L and to a lesser extent ss D S-L was not affected (Figure 5A). This suggests that the W/E residue can be efficiently removed from W/E S-L in the ER without the need for any other component. In order to further test this, precursors were transfected into HeLa-Kb cells that were treated with control or ERAP1-specific siRNA (Figure 5B). S-L presentation was reduced in ERAP1 knockdown cells transfected with ss D/W/E/M S-L as well as ss DD/WW/EE/MMS-L, demonstrating that ERAP1 was required for removal of the N-terminal amino acids in all of these precursors. Taken together this is suggestive of a mechanism which involves XXS-L retrotranslocation into the cytoplasm for XS-L generation followed by transport back into the ER for ERAP1 specific generation of S-L.
Figure 5. ER targeted precursors require cytosol trimming.
HeLa-Kb cells (black bars) and HeLa-Kb-ICP47 cells (white bars) were transfect with ss XS-L or ss XXS-L (A). Graph represents the average percentage presentation of two independent wells using the MFI of cells transfeted with ss MMS-L as 100% presentation. Error bars represent the difference within each group. (B) HeLa-Kb cells were treated with siRNA targeting ERAP1 (white bars) or ctrl siRNA (black bars) and transfected with ss XS-L or ss XXS-L. Graph represents the average percentage presentation of two independent experiments using the MFI of cells treated with ctrl siRNA and transfected with Ub MMS-L as 100% presentation. Error bars represent the difference within each group.
Discussion
Proteasomes generate many N-terminally-extended precursors. For these precursors to be presented on an MHC class I molecule they must be trimmed by peptidases to mature epitopes. Previous studies had shown that the removal of amino terminal flanking residues does indeed occur in living cells. This N-terminal trimming was “downstream” (independent) of proteasomes and mediated by aminopeptidases. It was clear that this peptide editing could in principle occur either in the cytosol, where peptides were first generated by proteasomes, and/or after transport into the ER by TAP. However, the relative contribution of these two compartments to trimming and their specificity had been incompletely understood.
Using fluorescent substrates and purified cytosol or ER the specificity of trimming in these compartments in vitro has been studied (31). However, in vivo the effect sequences N-terminal to an epitope have on processing and presentation on MHC class I molecules has only been systematically studied in the ER (32). This is important to define for cytosolic trimming because extracts may not faithfully reproduce the conditions in living cells (concentrations and ionic conditions are changed, enzymes may be activated or inactivated, metabolic pathways are inhibited, etc.) and presumably because of this the specificity of trimming we and Reits et al (9) observe in vivo is not identical to that reported by Shatz et al (31). Defining what is occurring in vivo is important biologically because the specificity of trimming can clearly influence the magnitude of responses and overall immunodominance hierarchies. Here we analyze the trimming of precursors in the cytosol of living cells and compare it to the trimming of the same precursors in the ER.
Our experimental approach was to express, in living cells, N-extended precursors in which we systematically varied the amino acids at the P2 and/or P1 position N-terminal to the SL8 epitope. Our findings clearly demonstrate that: (1) Trimming of these precursors can occur both in the cytosol and the ER; (2) The efficiency of cytosolic trimming process, like that of ER trimming (32) is affected by the N-terminal residues, i.e. it has specificity; (3) The specificity of cytosolic trimming is distinct from that in the ER; (4) Recycling of peptides from the ER to the cytosol may occur, potentially allowing sequential trimming of peptides in both compartments in either order; and (5) The net effect of cytosolic trimming is to broaden the repertoire of peptides that can be presented on MHC class I molecules.
Our experimental approach makes certain assumptions that are worth discussing. We expressed a series of peptides from minigenes that were transfected into antigen presenting cells. Our interpretation of the results assumes that the transcription, translation and, for Ub-X constructs, post-translational ubiquitin cleavage, are similar for all constructs. Because the ubiquitin construct bicistronically expresses GFP, we are able to gate on cells expressing similar levels of GFP that should also be expressing similar levels of the ubiquitin fusion proteins. To further test this assumption, we compared presentation from minigene constructs that are processed very differently and obtained very similar results using MAXXSL, Ub-XXS-L and Ub-XS-L constructs. This rules out the possibility that differences in presentation arising from differential ubiquitin-X cleavage and makes it highly unlikely that 1 (X) or 2 (XX) residues placed at different locations (2 or 76 residues) from the translational start site would affect translation (or transcription) and do so in the exact same way. Therefore, differences in translation, transcription and or Ub cleavage are unlikely to account for the differences in presentation that were seen with different specific sequences. While it is possible that these upstream residues may also affect TAP transport of cytosolic precursors, the residues that are consistently associated with high-level presentation of our model epitope are not particularly preferred for TAP translocation (40, 41). Moreover, we saw very similar results in mouse and human cells, although mouse and human TAP have somewhat different preferences for translocation (42–45). We interpret therefore, that presentation observed with the various epitope precursors reflects aminopeptidase specificity within the cell. However, it should be further pointed out that even if TAP selectivity contributes to some of the observed differences, our results still define the overall specificity of antigen presentation within the cell (i.e. cytosolic trimming, TAP transport and ER trimming). In addition, for the ER-targeted constructs we assume that XX residues do not influence cleavage by the signal peptidase (which liberates the epitope precursor from the signal peptide). This is supported by two pieces of data which show that the same results are obtained for sequences that are adjacent (ss X-S-L) or 6 residues away (ss LEQLXS-L) from the signal sequence cleavage site and that presentation from minigenes and trimming of the same sequences by purified ERAP1 correlates well with one another (32).
Our data show that much of the trimming of precursor peptides occurs in the ER, even when the peptides are originally generated in the cytosol, but the relative importance of ER vs. cytosolic trimming depends on the specific N-terminal residues. In one case, extensive trimming of a cytosolic precursor can occur in the cytosol (WWS-L). This N-terminal flanking sequence must be very efficiently removed in the cytosol, because when this same construct was targeted into the ER directly it was efficiently trimmed by ERAP1. In general, cytosolic trimming is more important for those sequences that are more poorly trimmed in the ER, broadening the sequences that can be efficiently processed and presented. As a result, the difference between the best and the worst presented constructs was smaller when N-extended peptides were expressed in the cytosol, compared to the ER. Only N-terminal glycine was associated with relatively poor presentation in both cytosolic and ER-targeted constructs. Nevertheless, the trimming that does occur in the cytosol does have specificity. There is a reproducible hierarchy, observed in both human and mouse cells, in the trimming and presentation of the various constructs based on the identity of their N-terminal residues.
Many of the precursors that were targeted by a signal sequence through SEC61 into the ER that were efficiently trimmed were presented equally well in control cells and cells in which TAP was inhibited with ICP47, as expected. However, a remarkable finding was that the presentation of some ER-targeted precursors was strongly inhibited by ICP47. It is formally possible that these were ones that failed to translocate through SEC61. However, this explanation seems unlikely because SEC61 can clearly transport the particular residues, when they are in other locations (e.g. E in S-L), or even when the residue is present as a singlet (ss XS-L) and SEC61 obviously transports proteins that have all 20 amino acids. Another possibility is that the signal peptidase failed to cleave the signal peptide from certain precursors, and these had to be retrotranslocated to the cytosol in order for the signal peptide to be removed. However, whether cleavage does occur depends mainly on features of the signal peptide that remain unchanged in all of the constructs used throughout this study, particularly the amino acids at positions −3 and −1 N-terminal of the cleavage site (reviewed in (46)). It seems more plausible that these sequences need export and trimming in the cytosol because they are very poorly trimmed in the ER. Indeed, the residues that are ICP47-inhibitable are the ones that are poorly removed by ERAP1 (32) and are particularly poorly removed when they are present in tandem.
Several previous studies have raised the possibility of export followed by re-import of peptides into the ER for presentation. For example, in vivo, epitopes are generated in a TAP-dependent mechanism from the signal sequence of LCMV gp33 protein (37), as are HLA-E-binding epitopes generated from the signal sequences of MHC class I molecules (38). Such TAP dependent presentation could be due to failed translocation of the signal sequence into the ER or retrotranslocation of peptide precursors from the ER into the cytosol. An elegant study by Altrich-VanLith et al has shown this to be the case for an epitope derived from tyrosinase (39). In this system, TAP inhibition significantly diminished presentation from ER targeted precursors with two-residue extensions containing histidine (HX). His is one of the amino acids poorly removed in the ER, and also required cytosolic trimming in our experimental system (ss HHS-L). In addition, earlier studies of isolated microsomes also demonstrated that ER-luminal peptides can be retrotranslocated out of the ER and recycle back in a TAP-dependent fashion (47). Thus there is growing evidence that ER-to-cytosol peptide translocation can occur, although the biological importance of this phenomenon was unknown. In these earlier studies why retrotranslocation might be important for presentation was not clear. Our findings suggest that ER-to-cytosol recycling may be important in situations where ERAP1 does not trim particular sequences down to mature epitopes. Our data further suggest that recycling of peptides between the ER and cytosol is likely to be a general phenomenon that occurs physiologically and contributes to MHC class I antigen presentation.
In conclusion our findings reveal that the amount of MHC-peptide complex presented to the immune system is determined by the identity of amino acids upstream of the epitope precursor and the ability of aminopeptidases to remove them. In many cases mature epitope generation is a combination of processing which has occurred in the cytosol and the ER. Taken together this demonstrates an important role of aminopeptidases in influencing the specificity of CD8+ T cell responses.
Supplementary Material
Acknowledgments
We thank Sopia Zendzian for technical assistance.
Abbreviations
- ER
endoplasmic reticulum
- ERAP1
Endoplasmic reticulum aminopeptidase 1
- MFI
mean fluorescence intensity
- MHC
major histocompatibility complex
- TAP
transporter associated with Ag processing
Footnotes
Disclosures
The authors have no financial conflict of interest.
This work was supported by grants from the National Institutes of Health (to K.L.R.) and core resources supported by the Diabetes Endocrinology Research Center Grant.
References
- 1.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL, Cascio P, Saric T, Rock KL. The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol Immunol. 2002;39:147–164. doi: 10.1016/s0161-5890(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 3.Rock KL, I, York A, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 4.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 5.Shastri N, Cardinaud S, Schwab SR, Serwold T, Kunisawa J. All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen-processing pathway. Immunol Rev. 2005;207:31–41. doi: 10.1111/j.0105-2896.2005.00321.x. [DOI] [PubMed] [Google Scholar]
- 6.Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J Biol Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 7.Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kloetzel PM. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol. 2004;5:661–669. doi: 10.1038/ni1090. [DOI] [PubMed] [Google Scholar]
- 9.Reits E, Neijssen J, Herberts C, Benckhuijsen W, Janssen L, Drijfhout JW, Neefjes J. A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity. 2004;20:495–506. doi: 10.1016/s1074-7613(04)00074-3. [DOI] [PubMed] [Google Scholar]
- 10.Seifert U, Maranon C, Shmueli A, Desoutter JF, Wesoloski L, Janek K, Henklein P, Diescher S, Andrieu M, de la Salle H, Weinschenk T, Schild H, Laderach D, Galy A, Haas G, Kloetzel PM, Reiss Y, Hosmalin A. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat Immunol. 2003;4:375–379. doi: 10.1038/ni905. [DOI] [PubMed] [Google Scholar]
- 11.York IA, Bhutani N, Zendzian S, Goldberg AL, Rock KL. Tripeptidyl Peptidase II Is the Major Peptidase Needed to Trim Long Antigenic Precursors, but Is Not Required for Most MHC Class I Antigen Presentation. J Immunol. 2006;177:1434–1443. doi: 10.4049/jimmunol.177.3.1434. [DOI] [PubMed] [Google Scholar]
- 12.Beninga J, Rock KL, Goldberg AL. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J Biol Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 13.Stoltze L, Schirle M, Schwarz G, Schroter C, Thompson MW, Hersh LB, Kalbacher H, Stevanovic S, Rammensee HG, Schild H. Two new proteases in the MHC class I processing pathway. Nat Immunol. 2000;1:413–418. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 14.York IA, Mo AX, Lemerise K, Zeng W, Shen Y, Abraham CR, Saric T, Goldberg AL, Rock KL. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity. 2003;18:429–440. doi: 10.1016/s1074-7613(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 15.Towne CF, I, York A, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL. Leucine aminopeptidase is not essential for trimming peptides in the cytosol or generating epitopes for MHC class I antigen presentation. J Immunol. 2005;175:6605–6614. doi: 10.4049/jimmunol.175.10.6605. [DOI] [PubMed] [Google Scholar]
- 16.Towne CF, I, York A, Watkin LB, Lazo JS, Rock KL. Analysis of the role of bleomycin hydrolase in antigen presentation and the generation of CD8 T cell responses. J Immunol. 2007;178:6923–6930. doi: 10.4049/jimmunol.178.11.6923. [DOI] [PubMed] [Google Scholar]
- 17.Towne CF, I, York A, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL. Puromycin-sensitive aminopeptidase limits MHC class I presentation in dendritic cells but does not affect CD8 T cell responses during viral infections. J Immunol. 2008;180:1704–1712. doi: 10.4049/jimmunol.180.3.1704. [DOI] [PubMed] [Google Scholar]
- 18.Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 19.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 20.Momburg F, Roelse J, Hammerling GJ, Neefjes JJ. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J Exp Med. 1994;179:1613–1623. doi: 10.1084/jem.179.5.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craiu A, Akopian T, Goldberg A, Rock KL. Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc Natl Acad Sci U S A. 1997;94:10850–10855. doi: 10.1073/pnas.94.20.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 23.Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL, Tsujimoto M, Goldberg AL. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 24.Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc Natl Acad Sci U S A. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity. 2006;25:795–806. doi: 10.1016/j.immuni.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci U S A. 2006;103:9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammer GE, Gonzalez F, Champsaur M, Cado D, Shastri N. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat Immunol. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 28.Yan J, V, Parekh V, Mendez-Fernandez Y, Olivares-Villagomez D, Dragovic S, Hill T, Roopenian DC, Joyce S, Van Kaer L. In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J Exp Med. 2006;203:647–659. doi: 10.1084/jem.20052271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammer GE, Gonzalez F, James E, Nolla H, Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat Immunol. 2006;8:101–108. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- 30.Blanchard N, Gonzalez F, Schaeffer M, Joncker NT, Cheng T, Shastri AJ, Robey EA, Shastri N. Immunodominant, protective response to the parasite Toxoplasma gondii requires antigen processing in the endoplasmic reticulum. Nat Immunol. 2008;9:937–944. doi: 10.1038/ni.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schatz MM, Peters B, Akkad N, Ullrich N, Martinez AN, Carroll O, Bulik S, Rammensee HG, van Endert P, Holzhutter HG, Tenzer S, Schild H. Characterizing the N-terminal processing motif of MHC class I ligands. J Immunol. 2008;180:3210–3217. doi: 10.4049/jimmunol.180.5.3210. [DOI] [PubMed] [Google Scholar]
- 32.Hearn A, York IA, Rock KL. The Specificity of Trimming of MHC Class I-Presented Peptides in the Endoplasmic Reticulum. J Immunol. 2009;183:5526–5536. doi: 10.4049/jimmunol.0803663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.York IA, Grant EP, Dahl AM, Rock KL. A mutant cell with a novel defect in MHC class I quality control. J Immunol. 2005;174:6839–6846. doi: 10.4049/jimmunol.174.11.6839. [DOI] [PubMed] [Google Scholar]
- 34.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 35.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 36.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 37.Hombach J, Pircher H, Tonegawa S, Zinkernagel RM. Strictly transporter of antigen presentation (TAP)-dependent presentation of an immunodominant cytotoxic T lymphocyte epitope in the signal sequence of a virus protein. J Exp Med. 1995;182:1615–1619. doi: 10.1084/jem.182.5.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braud VM, Allan DS, Wilson D, McMichael AJ. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol. 1998;8:1–10. doi: 10.1016/s0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 39.Altrich-VanLith ML, Ostankovitch M, Polefrone JM, Mosse CA, Shabanowitz J, Hunt DF, Engelhard VH. Processing of a class I-restricted epitope from tyrosinase requires peptide N-glycanase and the cooperative action of endoplasmic reticulum aminopeptidase 1 and cytosolic proteases. J Immunol. 2006;177:5440–5450. doi: 10.4049/jimmunol.177.8.5440. [DOI] [PubMed] [Google Scholar]
- 40.Peters B, Bulik S, Tampe R, Van Endert PM, Holzhutter HG. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol. 2003;171:1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
- 41.Doytchinova I, Hemsley S, Flower DR. Transporter associated with antigen processing preselection of peptides binding to the MHC: a bioinformatic evaluation. J Immunol. 2004;173:6813–6819. doi: 10.4049/jimmunol.173.11.6813. [DOI] [PubMed] [Google Scholar]
- 42.Momburg F, Neefjes JJ, Hammerling GJ. Peptide selection by MHC-encoded TAP transporters. Curr Opin Immunol. 1994;6:32–37. doi: 10.1016/0952-7915(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 43.Schumacher TN, Kantesaria DV, Heemels MT, Ashton-Rickardt PG, Shepherd JC, Fruh K, Yang Y, Peterson PA, Tonegawa S, Ploegh HL. Peptide length and sequence specificity of the mouse TAP1/TAP2 translocator. J Exp Med. 1994;179:533–540. doi: 10.1084/jem.179.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Endert PM, Riganelli D, Greco G, Fleischhauer K, Sidney J, Sette A, Bach JF. The peptide-binding motif for the human transporter associated with antigen processing. J Exp Med. 1995;182:1883–1895. doi: 10.1084/jem.182.6.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang P, Gyllner G, Kvist S. Selection and binding of peptides to human transporters associated with antigen processing and rat cim-a and -b. J Immunol. 1996;157:213–220. [PubMed] [Google Scholar]
- 46.Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 47.Roelse J, Gromme M, Momburg F, Hammerling G, Neefjes J. Trimming of TAP-translocated peptides in the endoplasmic reticulum and in the cytosol during recycling. J Exp Med. 1994;180:1591–1597. doi: 10.1084/jem.180.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.