Abstract
We previously reported that passive transfer of polyclonal neutralizing antibodies (NAbs) sufficient to generate a titer of 1:38 in the plasma would confer sterilizing protection to 99% of macaques challenged intravenously with 75 TCID50 of SHIVDH12. Neutralizing activity in that study was measured in an MT4 cell assay in which infection was completely blocked (EC100). In the current study, the TZM-bl system was used to measure EC50 neutralizing titers in several of the same macaque plasma samples and the relationship between these titers and in vivo protection was determined. The antiviral EC50 NAb titers measured in individual plasma samples were higher than those previously obtained in the MT4 system. Furthermore, the geometric mean EC50 NAb titers against pseudotyped SHIVDH12 were 33-fold greater than the EC100 titers measured in the MT4 cell assay against the replication-competent SHIVDH12 inoculated into animals. An augmented probit regression model was used to generate curves relating TZM-bl EC50 NAb titers and protection from a virus challenge; estimated titers conferring various levels of protection were then determined. In TZM-bl assays using pseudotyped SHIVDH12, representative percent in vivo protection/estimated EC50 titers were 99%/1:4467, 90%/1:1175, 80%/1:676, 50%/1:234, and 33%/1:141. Because it is likely that contributions from other arms of the immune system will contribute to vaccine-induced control, the range of EC50 NAb titers we have derived may be more informative for evaluating the protective value of NAb activity from TZM-bl assays.
Introduction
The development of an effective vaccine against the human immunodeficiency virus (HIV) is critical for controlling the acquired immunodeficiency syndrome (AIDS) epidemic. Nonhuman primates have been extensively used in conjunction with simian immunodeficiency virus (SIV) or the chimeric simian/human immunodeficiency virus (SHIV) in preclinical studies to monitor the effectiveness of new HIV-1 vaccine approaches. A variety of assays have been employed to measure the antiviral cellular and humoral responses elicited by these vaccines to delineate correlates of protection. The results from studies involving both SIVs and SHIVs have shown that prechallenge cellular responses alone do not prevent virus acquisition, although they can reduce the peak and set point levels of viremia.1–4 The contribution of vaccine-induced neutralizing antibodies (NAbs) in controlling virus infection has been more difficult to evaluate. Many vaccine regimens were either not designed to elicit or failed to generate NAbs against the challenge virus.1–9 In a few SHIV-based vaccine studies, the induction of vaccine-induced NAbs was associated with sterilizing protection, although a contribution of cellular immune responses could not be conclusively ruled out.10,11 The most unambiguous demonstration that preexisting NAbs can be protective comes from passive transfer studies in which the administration of neutralizing immunoglobulin (IgG) or neutralizing monoclonal antibodies prior to virus challenge conferred sterilizing protection.12–17
Whereas passive transfer experiments represent a “proof of concept” for NAb-mediated protection, a vaccine(s) capable of consistently eliciting such antibodies against genetically homologous or heterologous primate lentiviruses has not yet been developed. Several characteristics intrinsic to these viruses have made this a formidable challenge. First, NAbs invariably target the viral envelope (Env) protein, which exhibits a high degree of genetic and antigenic variability.18 A second major obstacle involves the emergence of new antigenic variants in an infected individual, which escape NAbs generated against viruses present at earlier times.19–21 Nonetheless, a few monoclonal NAbs, which target conserved regions within the viral Env, have been generated from clinical specimens collected from HIV-1-infected persons.22 Although these types of antibodies are rarely detected,23,24 and it is currently unknown how to elicit similar humoral responses, the identification of these and additional NAbs that target conserved Env epitopes is considered to be a critical step in developing immunogens that can counter the effects of antigenic variability and drift.
Measurements of the level and breadth of NAb responses in samples collected from vaccinees, infected individuals, or HIV surrogate animal models have relied on a variety of antiviral neutralization assays.17,20,21,25–30 Early assays were based on the ability of NAbs to completely block or suppress virus replication in peripheral blood mononuclear cells (PBMCs) or T cell line cultures.16,17,26,29,30 Because these assays relied on the control of spreading virus infections, cultures had to be maintained for extended periods of time for the antiviral neutralization effects to become evident. These labor-intensive systems were subsequently supplanted by newer high-throughput assays. Currently, the TZM-bl reporter cell system is the most widely used NAb assay.28 It utilizes replication-defective Env-pseudotyped challenge viruses produced from transfected 293T cells and the readout is inhibition of virus entry (e.g., 50% reduction in virus-induced luciferase activity or EC50). In studies assessing the breadth of antivirus inhibition, the TZM-bl assay has been employed to screen NAb activities in plasma samples from infected individuals that are directed against HIV isolates bearing genetically diverse viral Envs.23,25 Furthermore, because the same neutralization assay format is now used in many laboratories, a degree of standardization in the HIV-1 vaccine field has been achieved that allows the results from different laboratories to be compared.28 Although ever-increasing amounts of data have been collected with the TZM-bl system, little is known relating the NAb titers obtained to the protection of individuals or animals subsequently exposed to primate lentiviruses.
We have previously administered purified IgG containing high titered polyclonal NAbs against SHIVDH12 to 21 pigtailed macaques (Macaca nemestrina) and determined the NAb titer (1:38) in the plasma required to protect 99% of the animals against an intravenous (IV) challenge of 75 TCID50.15 This protective titer was derived using Reed–Muench analysis31 of the data from a 14-day spreading infection assay, which measures the complete inhibition of SHIVDH12 replication in cell cultures at the limiting dilution end point. The current study was undertaken to assess the protective NAb titers in several of the same animal plasma samples using the TZM-bl reporter system. Because the two assays used different readouts (complete inhibition of spreading infections and 50% reduction of virus entry), statistical approaches were developed to compare the NAb titers derived from both systems and to compile sets of deduced NAb titers conferring a range (33–99%) of protection against the SHIV infection in challenged animals.
Materials and Methods
Viruses stocks and titrations
The preparation of the original macaque PMBC-derived SHIVDH12 stock has been previously described.32 Replication-competent 293T-derived SHIVDH12 was generated by transfecting 293T cells with 7.5 μg of the full-length pSHIVDH12 plasmid molecular clone DNA32 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. The transfected cells were incubated for 48 h, virus-containing supernatants were collected, filtered through a 0.45-μm filter (Costar, Corning, NY), and aliquots were stored at −70°C. To generate 293T-derived SHIVDH12 pseudotyped virus, two separate plasmids were constructed. The first contained a −2 frameshift mutation that was introduced beginning at nucleotide 306 of the env gene of the full-length CCR5-tropic SHIVAD8 molecular clone (R. Willey and R. Sadjadpour, unpublished), creating the plasmid pSHIVAD8env−. A plasmid expressing the SHIVDH12 env gene, pCMVDHenv, was generated by polymerase chain reaction (PCR) amplification of the rev, vpu, and env genes from the pHIV-1DH12 molecular clone33 and subcloning into the NotI sites of the pCMVbeta expression plasmid (Clontech, Palo Alto, CA). Both plasmids (5 μg pSHIVAD8env− and 2 μg pCMVDHenv) were cotransfected into 293T cells and the resulting pseudotyped virus was collected and stored as described.
The PBMC and 293T cell-derived virus stocks were titrated by end point dilution in the TZM-bl cell line20, 21 and measuring luciferase activity expressed as relative luminescence units (RLU). Four-fold serial dilutions of the virus stocks were performed in complete Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS) and supplemented with 25 mM HEPES, 2 mM glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. Forty microliters from each virus dilution was added to each of four replicate 96-well flat bottom culture wells (Costar, Corning, NY), followed by the addition of 10 μl of heat-inactivated pigtailed macaque plasma (diluted 1:4). Replicate cultures without virus (mock infected) were also tested in parallel for each titration. Following a 1-h incubation at 37°C, 1 × 104 TZM-bl cells in 150 μl DMEM medium and DEAE dextran (20 μg/ml) were added to each well (DEAE dextran final concentration 15 μg/ml) and the cultures were incubated for an additional 28 h. The 28-h incubation time was selected to minimize additional rounds of virus replication. Cultures were incubated for 48 h when pseudotyped virus was titered. Following the virus incubation period, the medium was removed from each well by aspiration, the cells were lysed in 80 μl of luciferase cell lysis buffer (Promega, Madison, WI), and the RLU in 30 μl of each lysate was measured using a commercial luciferase detection kit (Promega, Madison, WI) and an LB940 Multilabel Reader (Berthold Technologies, Dresher, PA). The RLU was determined from each set of replicate wells and adjusted for background activity by subtracting the RLU measured in the mock-infected cultures. The amount of replication-competent PBMCs and 293T-dervied virus required to give 5 × 104 RLU per well was derived from titration curves. In the case of the pseudotyped virus stock, the amount of virus required to give 1 × 105 RLU was determined.
Virus neutralization assays
The collection and preparation of plasma samples from pigtailed macaques, 24 h following the administration of anti-HIV-1 neutralizing IgG, have been previously described.15,17 All samples, including plasma collected from a naive pigtailed macaque that had not received any IgG, were heated to 56°C for 1 h and insoluble material was removed by centrifugation prior to use. Each sample was initially diluted 1:4 in DMEM followed by 2-fold serial dilutions in 1:4-diluted naive pigtailed macaque plasma, so that all of the cultures assayed contained the same amount of total pigtailed macaque plasma (final dilution of 1:20 with virus; 1:80 after the addition of cells). Dilutions of the different virus stocks were prepared in DMEM so that 40 μl would generate the desired amount of RLU per well. Each titration also contained wells without virus for measurements of background luciferase activity. A total of 10 μl of each diluted plasma was added to the appropriate wells, the plates were incubated for 1 h at 37°C, and TZM-bl cells were added as described above. The lowest dilution for all plasma samples, mixed with virus prior to the addition of cells, was 1:20. Cultures were incubated for 28 or 48 h following the addition of cells for the replication-competent and pseudotyped viruses, respectively, and the RLU determined as described above. Plasma dose–response curves from the titrations were plotted and the plasma dilution causing a 50% reduction in the RLU, compared to virus with naive plasma controls, was determined as described below. Two independent titration experiments were performed for each virus and plasma sample.
Neutralization EC50 determinations and statistical analyses
EC50s were calculated from the TZM-bl assays using the four-parameter logistic model for each plasma sample and individual virus preparations; the EC50s from replicate assays were then averaged. If neither replica reached the 50% level, an EC50 value, which was half of the lowest observed value for any other plasma sample, was used. The relationship between the titers determined in the TZM-bl and MT4 assays was estimated using linear regression and Pearson's correlation.
Probit regression was used to model the relationship between the titers for complete suppression of virus replication obtained from the MT4 cell neutralization assay and in vivo protection using all 21 monkeys.34 Probit regression assumes that the probability of infection for a given MT4 readout = X is given by 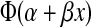 where α and β are estimated using maximum likelihood and Φ(z) is the standard normal cumulative distribution function evaluated at z. Titers for various levels of protection (e.g., 33%, 99%) were determined from the probit model estimates and the method of profile likelihood was used to construct 95% confidence intervals.35 Likelihood ratio tests were used to test whether a relationship (i.e., nonzero probit slope β) between the MT4 cell neutralization assay and in vivo protection existed.
where α and β are estimated using maximum likelihood and Φ(z) is the standard normal cumulative distribution function evaluated at z. Titers for various levels of protection (e.g., 33%, 99%) were determined from the probit model estimates and the method of profile likelihood was used to construct 95% confidence intervals.35 Likelihood ratio tests were used to test whether a relationship (i.e., nonzero probit slope β) between the MT4 cell neutralization assay and in vivo protection existed.
An augmented probit regression was used to relate TZM-bl readouts to in vivo protection. For the 11 macaques with experimentally measured neutralization titers, standard probit regression applied and the probability of in vivo protection given a TZM-bl readout = x is given by 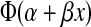 , as described above. For the 10 monkeys with no plasma samples, a range of plausible TZM-bl values, based on MT4 assay readouts and linear regression, was obtained. For those monkeys with just an MT4 assay readout = X, the probability of protection is given by a weighted average of
, as described above. For the 10 monkeys with no plasma samples, a range of plausible TZM-bl values, based on MT4 assay readouts and linear regression, was obtained. For those monkeys with just an MT4 assay readout = X, the probability of protection is given by a weighted average of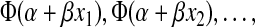 where the potential TZM-bl readouts x1, x2 … are weighted according to their probability given the MT4 = X. Intuitively, given a tight linear relationship between the MT4 and TZM-bl readouts, there will be a tight interval of plausible TZM-bl readouts. Importantly, this approach does not simply substitute the unobserved TZM-bl titer with its predicted value based on MT4 and linear regression, but takes into account the uncertainty of the prediction. As was the case for the MT4 assay, likelihood methods were used to estimate parameters, to obtain the ID33, ID99, etc., CIs, and to test for the existence of a relationship between neutralization titers and in vivo protection (see the Appendix).
where the potential TZM-bl readouts x1, x2 … are weighted according to their probability given the MT4 = X. Intuitively, given a tight linear relationship between the MT4 and TZM-bl readouts, there will be a tight interval of plausible TZM-bl readouts. Importantly, this approach does not simply substitute the unobserved TZM-bl titer with its predicted value based on MT4 and linear regression, but takes into account the uncertainty of the prediction. As was the case for the MT4 assay, likelihood methods were used to estimate parameters, to obtain the ID33, ID99, etc., CIs, and to test for the existence of a relationship between neutralization titers and in vivo protection (see the Appendix).
Results
Plasma NAb titers measured in the TZM-bl system
We previously reported that sterilizing protection of 99% of macaques inoculated intravenously with 75 TCID50 of SHIVDH12 required the passive transfer of antiviral IgG sufficient to generate a plasma NAb titer of 1:38 prior to virus exposure.15 This level of neutralizing activity was determined using Reed–Muench statistical analysis31 of a 14-day spreading infection neutralization assay in MT4 cells in which plasma samples from 21 IgG recipients were evaluated. In the current study, the antiviral titers of several of the same plasma samples were measured in the TZM-bl neutralization system to compare the protective in vivo NAb titers deduced from assays that differed both in format and readout. The neutralization sensitivities of three different SHIVDH12 preparations were evaluated using the TZM-bl assay (Table 1). Replication-competent SHIVDH12, prepared in macaque PBMCs and inoculated intravenously into the monkey recipients of the neutralizing IgG in the previous study, is designated SHIV PBMC. Infectious full-length SHIVDH12, produced in 293T cells following transfection with pSHIVDH12, is designated SHIV 293T. Replication-incompetent virus, pseudotyped with the SHIVDH12 Env, was generated in 293T cells as described in Materials and Methods and is designated SHIV (Pseudo) 293T. These three SHIV preparations were chosen to evaluate (1) producer cell effects (SHIV PBMC vs. SHIV 293T) and (2) pseudotyping [SHIV (Pseudo) 293 vs. SHIV 293T] on the neutralization titers obtained in the TZM-bl system.
Table 1.
SHIVs Used in Neutralization Assays
| Virus | Origin | Properties |
|---|---|---|
| SHIV PBMC | Infected macaque PBMCs | Replication-competent SHIVDH12 inoculum used in macaque studies |
| SHIV 293T | Transfected 293T cells | Replication-competent SHIVDH12 used in tissue culture experiments |
| SHIV (Pseudo) 293T | Transfected 293T cells | Pseudotyped replication incompetent virus bearing SHIVDH12 envelope |
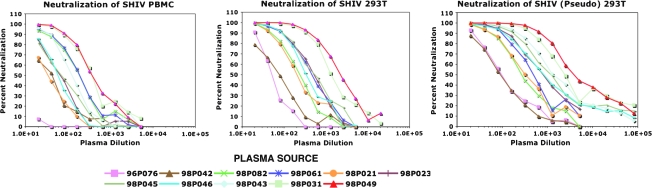
Plasma samples, collected 24 h following passive transfer, were available for testing from 11 of the original 21 animals and the dilution capable of reducing virus infectivity by 50% in the TZM-bl assay was determined. The titrations from one of two replicate neutralization assays for all of the plasma samples tested and for the three SHIV preparations are shown in Fig. 1. Visual inspection of the titration curves indicated that with one exception, all of the plasma samples had NAb activity sufficient to reduce the infectivity of all three viruses in TZM-bl cells by more than 50%. A 50% reduction was not attained with the 96P076 plasma sample against the SHIV PBMCs in either replicate assay at the lowest dilution (1:20) of plasma tested (Fig. 1, top panel). The NAb inhibition curves exhibited a general shift toward the higher plasma dilutions when the virus tested was produced in 293T cells, versus PBMCs, indicating that the SHIV 293T and SHIV (Pseudo) 293T stocks were more sensitive to NAbs than the SHIV PBMCs.
FIG. 1.
Neutralization of replication-competent and pseudotyped SHIVDH12 preparations with pigtailed macaque plasma specimens. Plasma samples were collected from the indicated animals 24 h following transfer of neutralizing IgG and immediately prior to SHIVDH12 challenge. The Nab activity was titrated using the TZM-bl assay.
Based on the data shown in Fig. 1, EC50 neutralization titers were calculated for the three viruses against individual monkey plasma samples using the four-parameter logistic model described in Materials and Methods. The average EC50 titers, determined from replicate TZM-bl assays, were then compared to the NAb titers previously determined in the MT4 cell system (Table 2, Observed). An increase in the TZM-bl-derived EC50 NAb titers was observed for all of the monkey plasma samples compared to those reported for SHIV PBMCs in our previous study, with the exception of 96P076 mentioned earlier. The geometric mean (gm) of the NAb titers measured against SHIV PBMCs in the TZM-bl system was 4.3-fold higher than that obtained using the MT4 cell assay. This increase was not unexpected as the TZM-bl readout relied on a 50% reduction (EC50) in virus entry versus the complete block [100% neutralization (EC100)] of spreading virus infections in MT4 cells. Furthermore, the gm NAb titers measured for SHIV 293T and SHIV (Pseudo) 293T in the TZM-bl assay were 20.6- and 33.1-fold higher, respectively, compared to the NAb titers previously measured in MT4 cells using SHIV PBMCs. Thus, both the cell of origin and the use of pseudotyped virus affected neutralization sensitivity in the TZM-bl system.
Table 2.
Comparisons of NAb Titers
| |
|
NAb assay in MT4 cells |
NAb assay in TZM-bl cells |
||
|---|---|---|---|---|---|
|
Observed |
|
SHIV PBMCs |
SHIV PBMCs |
SHIV 293T |
SHIV (Pseudo) 293T |
| Animal ID | Sterilizing protection | NAb titer in plasma (complete inhibition of spreading infection) | NAb titer in plasma EC50 (SD) | NAb titer in plasma EC50 (SD) | NAb titer in plasma EC50 (SD) |
| 98P049 | Yes | 1:123 | 1:332 (65) | 1:2531 (580) | 1:4074 (505) |
| 98P031 | Yes | 1:123 | 1:413 (125) | 1:1481 (117) | 1:1812 (529) |
| 98P043 | Yes | 1:40 | 1:117 (3) | 1:820a | 1:1484 (11) |
| 98P045 | Yes | 1:18 | 1:52 (24) | 1:428 (94) | 1:1015 (41) |
| 98P046 | No | 1:18 | 1:66 (12) | 1:371 (16) | 1:843 (14) |
| 98P023 | Yes | 1:15 | 1:54 (12) | 1:430 (66) | 1:854 (117) |
| 98P021 | Yes | 1:13 | 1:36 (25) | 1:201 (8) | 1:375 (93) |
| 98P061 | Yes | 1:12 | 1:195 (23) | 1:392 (76) | 1:417 (48) |
| 98P082 | No | 1:7 | 1:177 (5) | 1:275 (81) | 1:278 (10) |
| 98P042 | Yes | 1:6 | 1:30 (9) | 1:68 (11) | 1:92a |
| 96P076 | No | 1:3 | 1:5b | 1:55 (7) | 1:96 (1) |
|
Predicted |
|
SHIV PBMCs |
SHIV PBMCs |
SHIV 293T |
SHIV (Pseudo) 293T |
|---|---|---|---|---|---|
| Animal ID | Sterilizing protection | NAb titer in plasma (complete inhibition of spreading infection) | NAb titer in plasma (80% prediction interval for EC50)c | NAb titer in plasma (80% prediction interval for EC50) | NAb titer in plasma (80% prediction interval for EC50) |
| 98P024 | Yes | 1:40 | 1:55–1:420 | 1:480–1:1272 | 1:1717–1:2230 |
| 94P011 | Yes | 1:8 | 1:14–1:109 | 1:107–1:284 | 1:158–1:491 |
| 98P035 | No | 1:7 | 1:13–1:97 | 1:95–1:251 | 1:139–1:433 |
| 94P040 | Yes | 1:5 | 1:10–1:73 | 1:69–1:184 | 1:101–1:315 |
| 94P028 | No | 1:4 | 1:18–1:61 | 1:56–1:149 | 1:82–1:256 |
| 96P088 | Yes | 1:4 | 1:8–1:61 | 1:56–1:149 | 1:82–1:256 |
| 96P081 | No | 1:4 | 1:8–1:61 | 1:56–1:149 | 1:82–1:256 |
| 94P032 | No | 1:3 | 1:6–1:48 | 1:34–1:114 | 1:63–1:195 |
| 94P029 | No | 1:3 | 1:6–1:48 | 1:34–1:114 | 1:63–1:195 |
| 94P027 | No | 1:3 | 1:6–1:48 | 1:34–1:114 | 1:63–1:195 |
An EC50 neutralization titer was obtained from only one replicate.
Estimated EC50; a 50% neutralization titer could not be obtained for either replicate.
80% predicted TZM-bl NAb titer intervals based on a linear regression model for animals for which plasma was no longer available (see Fig. 3).
Determining in vivo protection titers
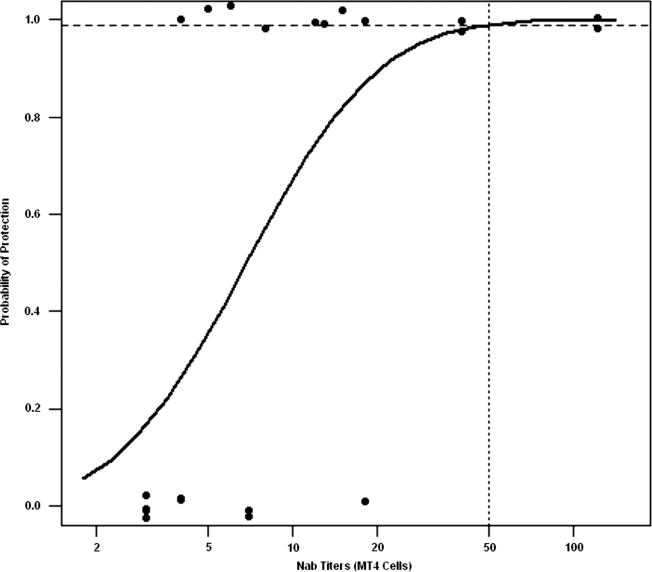
As noted earlier, Reed–Muench analysis of data from our previous study, in which NAb titers were defined as the highest dilution of plasma preventing virus infection in MT4 cell cultures, predicted that a neutralizing titer of 1:38 would confer 99% protection to macaques challenged intravenously with 75 TCID50 of SHIVDH12. When probit regression was used to analyze the neutralization results for the 21 macaques evaluated in the previous study, a significant relationship between the MT4 cell-derived titers and the probability of infection was observed (p = 0.02). Based on the probit model, an NAb titer of 1:50 (95% CI 1:25, 1:4300) was required to confer 99% protection (Fig. 2).
FIG. 2.
Probit regression analysis relating the NAb titers determined in the MT4 cell assay and protection from SHIVDH12 infection. Titers for different levels of protection for 21 macaques were determined from probit model estimates and profile likelihood was used to construct 95% confidence intervals as described in Materials and Methods and in the Appendix. The NAb titer estimated to protect 99% of challenged animals is indicated by the vertical dotted line.
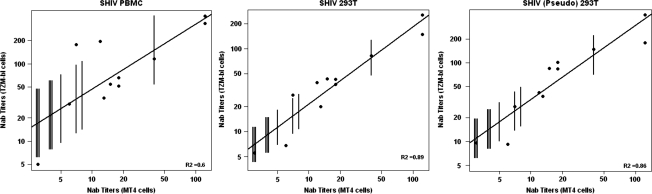
A major goal of this study was to relate EC50 levels, determined in the TZM-bl system, to neutralizing titers conferring protection to macaques challenged with SHIVDH12 and to include plasma samples from all 21 macaques, previously evaluated using the MT4 cell neutralization assay, in the analysis. As noted earlier, plasma specimens from only 11 of the original animals were available. To obtain EC50 values for the 10 monkeys with no plasma samples available for testing in the TZM-bl system, linear regression was used to compare the neutralization titers obtained against all three SHIV preparations in both assays. Eighty percent prediction intervals for the EC50 values of these 10 animals were determined from these regression curves (Fig. 3, Table 2 predicted).
FIG. 3.
Linear regression analyses comparing NAb titers determined in the TZM-bl cell and MT4 cell assays. The black dots indicate the experimentally derived data using available plasma samples from 11 macaques. The vertical lines indicate 80% prediction intervals for EC50 values for the 10 animals from which no plasma samples were available as described in Materials and Methods. The three SHIV preparations used for neutralization assays are indicated.
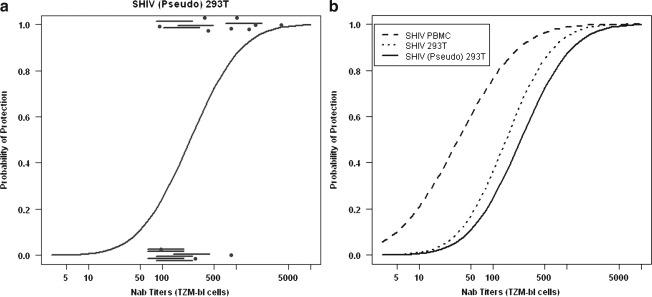
To estimate the relationship between the EC50 neutralization titers for all 21 macaques and protection from a subsequent SHIV infection, the augmented probit regression model described in Materials and Methods was used. Figure 4a shows the estimated probit fitted curve for SHIV (Pseudo) 293T; the experimentally determined results plus the predicted data with the 80% intervals are each indicated. The fitted curves for all three SHIV preparations relating protection in vivo with TZM-bl neutralization titers are presented in Fig. 4b. Estimated titers, conferring various levels of protection, were derived from these probit curves and are shown in Table 3 along with the p-value for the test of a relationship between NAb titer and protection. Taken together, the analyses for all three viruses show a significant relationship between EC50 titers and protection from infection (p = 0.02 or 0.01).
FIG. 4.
Augmented probit regression analyses relating EC50 NAb titers determined using the TZM-bl system and protection from SHIVDH12 infection. (a) The 11 dots are the experimentally derived EC50 titers from the 11 macaques from which plasma samples were available. The horizontal lines represent predicted 80% confidence intervals for EC50 titers for the 10 animals from which plasma specimens were no longer available. NAb titers for different levels of protection were determined as described in Materials and Methods and the Appendix and a fitted curve for this data set is shown. (b) Fitted curves from augmented probit analyses relating NAb titers, determined in the TZM-bl system against the indicated SHIVs, and in vivo protection are shown.
Table 3.
Animal Protective Neutralizing Antibody Titers
| |
|
|
|
% Protection |
||||
|---|---|---|---|---|---|---|---|---|
| Assay | Virus | Animals (n) | p-Value | 99 | 90 | 80 | 50 | 33 |
| MT4 cell | SHIV PBMCs | 21 | 0.02 | 50 (24.7, 4.303)a | 20 (11, 249) | 14 (8, 79) | 7 (3, 14) | 5 (1, 8) |
| TZM-bl | SHIV PBMCs | 21 | 0.02 | 1096 (174, 2.8211) | 229 (73, 5.776) | 118 (45, 6.804) | 34 (2.5, 107) | 18 (<1, 46) |
| TZM-bl | SHIV 293T | 21 | 0.007 | 2204 (587, 7.126) | 661 (263, 5.144) | 398 (185, 6.983) | 151 (40, 365) | 93 (8, 192) |
| TZM-bl | SHIV (Pseudo) 293T | 21 | 0.009 | 4467 (978, 4.977) | 1175 (438, 1.775) | 676 (299, 1794) | 234 (53, 618) | 141 (8, 308) |
The NAb titers calculated from the augmented probit analyses shown in Figs. 2 and 4b are indicated in bold.
95% confidence interval.
The NAb titers for SHIV PBMCs provide a direct comparison of the two assay formats to predict protection against a SHIVDH12 infection. As noted above, probit regression estimated that a titer of 1:50, against the SHIV PBMCs in the MT4 assay, would protect 99% of the challenged animals. When the same virus and plasma samples were evaluated in the TZM-bl system, 99% protection in vivo required a titer of 1:1096 (95% CI 1:174, 1:2 × 1011). If SHIV (Pseudo) 293T was the test virus evaluated in the TZM-bl assay, the calculated 99% titer increased to 1:4467 (95% CI 1:978, 5.0 × 107). A 99% protective NAb titer may be too difficult to achieve with any vaccine regimen or may not be required in lieu of contributions by other components of the immune system. Therefore, the lower protection estimates and corresponding NAb titers shown in Table 3 may provide for a more realistic evaluation of the results derived from the TZM-bl analyses. In this regard, more modest NAb titers (1:234; CI 1:53 to 1:618) would be required to achieve 50% protection in vivo when pseudotyped virus is used to screen NAb in plasma samples using the TZM-bl assay. It is also important to note that the estimated protective NAb titers shown for the SHIV (Pseudo) 293T are potentially the most applicable to current studies assessing NAb activity since pseudotyped HIV-1, SIV, and SHIV preparations are the most commonly used viruses in TZM-bl assays to measure plasma NAb levels in preclinical vaccine experiments.
Discussion
The development and use of engineered cell lines, in combination with Env-pseudotyped viruses able to activate reporter genes,20,21 have greatly simplified measurements of NAbs present in HIV-1-infected individuals and in SIV- and SHIV-infected animals and have reduced the interlaboratory variability in monitoring neutralizing activity.28 The TZM-bl cell-based assay has standardized and facilitated high-throughput screenings for broadly reacting neutralizing activities23,25,36,37 and studies of the emergence of neutralization escape variants in HIV-infected individuals.20,21,38 Despite the plethora of anti-HIV and SIV/SHIV NAb data generated with the TZM-bl system, the assay has yet to yield information relating the virus neutralization titers obtained to in vivo NAb levels that confer protection to individuals or animals exposed to virus.
We and others have previously reported the levels of NAbs required for protection against virus challenge in the X4-tropic SHIV macaque animal model following the transfer of anti-HIV-1 neutralizing IgG or monoclonal antibodies.12–17 In earlier experiments, we used a 14-day MT4 cell-based neutralization assay and reported that a plasma NAb titer of 1:38 at the time of virus challenge would protect 99% of the animals against SHIVDH12 infection, as determined by Reed–Muench analysis.15 In the current study, the same data were evaluated using standard probit regression. An advantage of probit regression is its ability to provide confidence intervals that reflect the uncertainty based on smaller sample sizes (21 monkeys). Probit regression analysis estimated that a plasma NAb titer of 1:50 (95% CI 1:25, 1:4300) determined in the MT4 cell assay would protect 99% of virus-challenged animals. This titer is similar to that (viz. 1:38) predicted by Reed–Muench,31 although there is uncertainty about the actual value as reflected in the confidence interval.
The major aim of the current study was to determine the relationship between neutralization titers determined in the TZM-bl assay and protection in the SHIV/macaque model. The strong linear relationship between the neutralization titers obtained from the TZM-bl and MT4 cell assays (Fig. 3) allowed us to incorporate data from all 21 monkeys from the original study and link the TZM-bl EC50 results with the subsequent protection/infection of SHIV-inoculated animals. It should be noted that the confidence intervals derived from the augmented probit model incorporate the uncertainty in the estimates arising from the sample of 21 monkeys as well as the additional uncertainty involving approximating the TZM-bl titers for 10 of these animals.
Our results indicate that when neutralization of SHIV PBMC was assessed in the TZM-bl system, a titer of 1:1096 was estimated to confer 99% protection against a SHIVDH12 challenge (Table 3). This NAb titer is significantly higher than the protective titer (1:50) determined by probit regression analysis of the data derived from the previously reported MT4 cell neutralization assay. When SHIV (Pseudo) 293T was used as the test virus in the TZM-bl system, the plasma NAb titer calculated to confer 99% protection increased to 1:4467. The titers determined in the MT4 cell and TZM-bl assays are clearly based on different readouts making a direct comparison difficult. Furthermore, the use of the more sensitive 293T cell-derived and pseudotyped viruses both contributed to protective neutralizing titers that were much higher than those obtained from the MT4 cell assay using SHIV PBMCs. The influences of cell of origin and pseudotyping on the NAb titers are consistent with other studies, which reported that virus produced from 293 T cells was more sensitive to neutralization than genetically identical preparations generated in PBMCs and that pseudotyped virus was more sensitive to neutralization than replication-competent virus produced from 293 T cells.27,39 However, depending on the source of Nab tested, target cells used in the assay may also influence Nab titers obtained.40 Nonetheless, the range of protective neutralizing titers shown in Table 3 for SHIV (Pseudo) 293T may be most relevant when interpreting NAb efficacy in view of the widespread use of pseudotyped rather than replication-competent virus in the TZM-bl assay.
Protection mediated by antiviral NAbs can be defined in several ways when considered in the context of primate lentivirus infections. This includes the suppression of virus replication/set point viremia, the prevention/delay of disease, or the conferral of sterilizing immunity. In our experiments, protection has been defined as blocking the establishment of a SHIVDH12 infection in 99% of inoculated macaques, subsequent to the transfer of neutralizing IgG. This is an extremely stringent definition of NAb-mediated protection when applied to the possible beneficial effects derived from prophylactic vaccines, which may elicit multiple types of immune responses in addition to the production of NAbs. For example, previous reports have described anti-SIV/SHIV vaccine regimens that elicit cytotoxic T lymphocyte responses that are capable of reducing peak viremia and set point virus loads.1–3,6,8,9 Furthermore, antibody-dependent cell-mediated cytotoxicity activity (ADCC), observed during both HIV and SIV in vivo infections and associated with reduced plasma viral loads and slower disease progression,41–43 is not measurable in standard NAb assays, and could contribute to the “protection” seen following passive transfer of neutralizing IgG, as previously reported.44 Therefore, the range of NAb titers shown in Table 3 should be viewed in the context of these antibodies acting in concert with other vaccine-induced immune responses. It is worth noting that the range of neutralization activities shown was based on protecting animals from an intravenous SHIVDH12 challenge of 75 TCID50. It is likely that lower NAb titers might be required to protect macaques against smaller virus inocula or via a mucosal rather than an intravenous route.
It is also interesting to compare the protective neutralization titers shown in Table 3 for SHIV (Pseudo) 293T in the TZM-bl assay with published NAb titers measured in HIV-1-infected individuals using pseudotyped virus in the same assay. One study, assessing the neutralizing activities present in pooled sera from HIV-1 clade A- through D-infected individuals and directed against pseudotyped viruses bearing “early” envelope glycoproteins of diverse geographic origin, reported geometric mean NAb titers ranging from 1:97 to 1:361 in individual pools.27 Another study, comparing levels of plasma NAbs in HIV-superinfected individuals with patients carrying a single virus strain, reported that both cohorts had TZM-bl EC50 titers of approximately 1:100 and noted that this level of neutralizing activity was insufficient to block superinfection.45 Finally, in a passive transfer study involving the administration of a mixture of neutralizing monoclonal antibodies (2G12, 2F5, and 4E10) to HIV-1-infected individuals undergoing interruption of antiretroviral therapy, a plasma NAb titer of >1:200 was calculated to suppress virus rebound in 50% of responding patients.46 Although this latter report differed markedly from our passive transfer experiment (HIV-1 vs. SHIV; humans vs. macaques; suppression of a preexisting infection vs. control of an acute infection; EC70 vs. mean NAb EC50 titer determinations in the TZM-bl assay), the plasma NAb titers predicted to suppress viremia or achieve protection in 50% of patients or monkeys, respectively, were surprisingly similar (1:200 vs. 1:234). Our results have related the NAb titers deduced from TZM-bl assays and the control of lentivirus replication in vivo. However, the relationship of these titers to protection of humans following exposure to HIV-1 remains to be determined.
Appendix
In this appendix, we provide details on maximum likelihood estimation and confidence interval construction using the probit and augmented probit models. Let X be the log10 titer using the MT4 cell assay. The probit model assumes that the probability a monkey becomes infected is given by 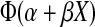 , where Φ(z) is the standard normal cumulative distribution function evaluated at z and α,β are unknown parameters to be estimated. Let y = 1 if a monkey is infected and 0 otherwise. We estimate the unknown parameters by maximizing the log-likelihood:
, where Φ(z) is the standard normal cumulative distribution function evaluated at z and α,β are unknown parameters to be estimated. Let y = 1 if a monkey is infected and 0 otherwise. We estimate the unknown parameters by maximizing the log-likelihood:
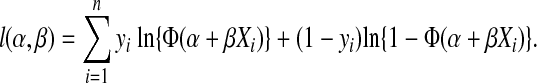 |
To construct 95% profile likelihood confidence intervals for the 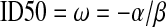 , we form the profile log-likelihood,
, we form the profile log-likelihood, 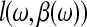 , where
, where 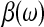 is the maximum likelihood estimate of β for a fixed ω. See Cox and Hinkley35 for details. Confidence intervals for the ID99 and ID33 etc. are similarly constructed. A likelihood ratio test is used to test whether β is 0.
is the maximum likelihood estimate of β for a fixed ω. See Cox and Hinkley35 for details. Confidence intervals for the ID99 and ID33 etc. are similarly constructed. A likelihood ratio test is used to test whether β is 0.
For the TMZ-bl assay, 10 monkeys have missing readouts but were evaluated in the MT4 cell assay. Let x be the log10 EC50 based on the TMZ-bl assay. For monkeys with x missing we assume that there is a linear relationship between the MT4 assay log10 titer X and the TMZ-bl log10 EC50 x:
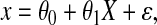 |
where ɛ has a Gaussian distribution with mean 0 and variance 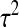 . The parameters
. The parameters 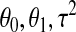 are estimated using least squares and treated as fixed. Under the above assumptions, it can be shown that for a monkey with an MT4 cell assay value of X, the probability of infection is given by
are estimated using least squares and treated as fixed. Under the above assumptions, it can be shown that for a monkey with an MT4 cell assay value of X, the probability of infection is given by
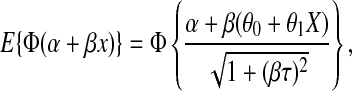 |
where the expectation E is with respect to the (Gaussian) distribution of x, which has a mean 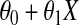 and variance
and variance 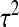 . We construct the augmented probit log-likelihood by using the above expression for the probability of infection in the probit log-likelihood for the 10 monkeys with unknown x. For the 11 monkeys for which x is measured, we use
. We construct the augmented probit log-likelihood by using the above expression for the probability of infection in the probit log-likelihood for the 10 monkeys with unknown x. For the 11 monkeys for which x is measured, we use 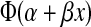 . Estimates of α,β are obtained by maximizing the augmented probit log-likelihood. Profile likelihood and likelihood ratio tests were used to construct confidence intervals and test if β were 0.
. Estimates of α,β are obtained by maximizing the augmented probit log-likelihood. Profile likelihood and likelihood ratio tests were used to construct confidence intervals and test if β were 0.
Acknowledgments
The authors acknowledge the John Mascola Laboratory and Tatsuhiko Igarashi for help in establishing the TZM-bl neutralization assay. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Disclosure Statement
No competing financial interests exist.
References
- 1.Amara RR. Villinger F. Altman JD, et al. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science. 2001;292(5514):69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- 2.Barouch DH. Santra S. Schmitz JE, et al. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290(5491):486–492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 3.Letvin NL. Mascola JR. Sun Y, et al. Preserved CD4 + central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312(5779):1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson NA. Reed J. Napoe GS, et al. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J Virol. 2006;80(12):5875–5885. doi: 10.1128/JVI.00171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casimiro DR. Wang F. Schleif WA, et al. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with dna and recombinant adenoviral vaccine vectors expressing Gag. J Virol. 2005;79(24):15547–15555. doi: 10.1128/JVI.79.24.15547-15555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J. O'Brien KL. Lynch DM, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457(7225):87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ourmanov I. Brown CR. Moss B, et al. Comparative efficacy of recombinant modified vaccinia virus Ankara expressing simian immunodeficiency virus (SIV) Gag-Pol and/or Env in macaques challenged with pathogenic SIV. J Virol. 2000;74(6):2740–2751. doi: 10.1128/jvi.74.6.2740-2751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz JE. Kuroda MJ. Santra S, et al. Control of viremia in simian immunodeficiency virus infection by CD8 + lymphocytes. Science. 1999;283(5403):857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 9.Shiver JW. Fu TM. Chen L, et al. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature. 2002;415(6869):331–335. doi: 10.1038/415331a. [DOI] [PubMed] [Google Scholar]
- 10.Barnett SW. Srivastava IK. Kan E, et al. Protection of macaques against vaginal SHIV challenge by systemic or mucosal and systemic vaccinations with HIV-envelope. AIDS. 2008;22(3):339–348. doi: 10.1097/QAD.0b013e3282f3ca57. [DOI] [PubMed] [Google Scholar]
- 11.Cho MW. Kim YB. Lee MK, et al. Polyvalent envelope glycoprotein vaccine elicits a broader neutralizing antibody response but is unable to provide sterilizing protection against heterologous simian/human immunodeficiency virus infection in pigtailed macaques. J Virol. 2001;75(5):2224–2234. doi: 10.1128/JVI.75.5.2224-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baba TW. Liska V. Hofmann-Lehmann R, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6(2):200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 13.Mascola JR. Lewis MG. Stiegler G, et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73(5):4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mascola JR. Stiegler G. VanCott TC, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6(2):207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura Y. Igarashi T. Haigwood N, et al. Determination of a statistically valid neutralization titer in plasma that confers protection against simian-human immunodeficiency virus challenge following passive transfer of high-titered neutralizing antibodies. J Virol. 2002;76(5):2123–2130. doi: 10.1128/jvi.76.5.2123-2130.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parren PW. Marx PA. Hessell AJ, et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J Virol. 2001;75(17):8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata R. Igarashi T. Haigwood N, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5(2):204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson-Hedestam GB. Fouchier RA. Phogat S. Burton DR. Sodroski J. Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6(2):143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 19.Frost SD. Wrin T. Smith DM, et al. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci USA. 2005;102(51):18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richman DD. Wrin T. Little SJ. Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100(7):4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei X. Decker JM. Wang S, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 22.Burton DR. Desrosiers RC. Doms RW, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5(3):233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 23.Li Y. Migueles SA. Welcher B, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13(9):1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheid JF. Mouquet H. Feldhahn N, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 25.Brown BK. Wieczorek L. Sanders-Buell E, et al. Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology. 2008;375(2):529–538. doi: 10.1016/j.virol.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 26.Burton DR. Pyati J. Koduri R, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 27.Li M. Gao F. Mascola JR, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polonis VR. Brown BK. Rosa Borges A, et al. Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination. Virology. 2008;375(2):315–320. doi: 10.1016/j.virol.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Willey RL. Shibata R. Freed EO. Cho MW. Martin MA. Differential glycosylation, virion incorporation, and sensitivity to neutralizing antibodies of human immunodeficiency virus type 1 envelope produced from infected primary T-lymphocyte and macrophage cultures. J Virol. 1996;70(9):6431–6436. doi: 10.1128/jvi.70.9.6431-6436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyand MS. Manson KH. Garcia-Moll M. Montefiori D. Desrosiers RC. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70(6):3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed LJ. Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 32.Shibata R. Maldarelli F. Siemon C, et al. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: Determinants of high virus loads and CD4 cell killing. J Infect Dis. 1997;176(2):362–373. doi: 10.1086/514053. [DOI] [PubMed] [Google Scholar]
- 33.Shibata R. Hoggan MD. Broscius C, et al. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69(7):4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finney DJ. Probit Analysis. 3d. Cambridge University Press; Cambridge, England: 2007. [Google Scholar]
- 35.Cox DR. Hinkley DV. Theoretical Statistics. Chapman & Hall; London, England: 1974. [Google Scholar]
- 36.Binley JM. Lybarger EA. Crooks ET, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82(23):11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraft Z. Derby NR. McCaffrey RA, et al. Macaques infected with a CCR5-tropic simian/human immunodeficiency virus (SHIV) develop broadly reactive anti-HIV neutralizing antibodies. J Virol. 2007;81(12):6402–6411. doi: 10.1128/JVI.00424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morner A. Douagi I. Forsell MN, et al. Human immunodeficiency virus type 1 env trimer immunization of macaques and impact of priming with viral vector or stabilized core protein. J Virol. 2009;83(2):540–551. doi: 10.1128/JVI.01102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Louder MK. Sambor A. Chertova E, et al. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology. 2005;339(2):226–238. doi: 10.1016/j.virol.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Mann AM. Rusert P. Berlinger L. Kuster H. Gunthard HF. Trkola A. HIV sensitivity to neutralization is determined by target and virus producer cell properties. AIDS. 2009;23(13):1659–1667. doi: 10.1097/QAD.0b013e32832e9408. [DOI] [PubMed] [Google Scholar]
- 41.Forthal DN. Landucci G. Daar ES. Antibody from patients with acute human immunodeficiency virus (HIV) infection inhibits primary strains of HIV type 1 in the presence of natural-killer effector cells. J Virol. 2001;75(15):6953–6961. doi: 10.1128/JVI.75.15.6953-6961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forthal DN. Landucci G. Haubrich R, et al. Antibody-dependent cellular cytotoxicity independently predicts survival in severely immunocompromised human immunodeficiency virus-infected patients. J Infect Dis. 1999;180(4):1338–1341. doi: 10.1086/314988. [DOI] [PubMed] [Google Scholar]
- 43.Gomez-Roman VR. Patterson LJ. Venzon D, et al. Vaccine-elicited antibodies mediate antibody-dependent cellular cytotoxicity correlated with significantly reduced acute viremia in rhesus macaques challenged with SIVmac251. J Immunol. 2005;174(4):2185–2189. doi: 10.4049/jimmunol.174.4.2185. [DOI] [PubMed] [Google Scholar]
- 44.Hessell AJ. Hangartner L. Hunter M, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449(7158):101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 45.Blish CA. Dogan OC. Derby NR, et al. Human immunodeficiency virus type 1 superinfection occurs despite relatively robust neutralizing antibody responses. J Virol. 2008;82(24):12094–12103. doi: 10.1128/JVI.01730-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trkola A. Kuster H. Rusert P, et al. In vivo efficacy of human immunodeficiency virus neutralizing antibodies: Estimates for protective titers. J Virol. 2008;82(3):1591–1599. doi: 10.1128/JVI.01792-07. [DOI] [PMC free article] [PubMed] [Google Scholar]