Abstract
In mid September 2008, clinical signs of bluetongue (particularly coronitis) were observed in cows on three different farms in eastern Netherlands (Luttenberg, Heeten, and Barchem), two of which had been vaccinated with an inactivated BTV-8 vaccine (during May-June 2008). Bluetongue virus (BTV) infection was also detected on a fourth farm (Oldenzaal) in the same area while testing for export. BTV RNA was subsequently identified by real time RT-PCR targeting genome-segment (Seg-) 10, in blood samples from each farm. The virus was isolated from the Heeten sample (IAH “dsRNA virus reference collection” [dsRNA-VRC] isolate number NET2008/05) and typed as BTV-6 by RT-PCR targeting Seg-2. Sequencing confirmed the virus type, showing an identical Seg-2 sequence to that of the South African BTV-6 live-vaccine-strain. Although most of the other genome segments also showed very high levels of identity to the BTV-6 vaccine (99.7 to 100%), Seg-10 showed greatest identity (98.4%) to the BTV-2 vaccine (RSAvvv2/02), indicating that NET2008/05 had acquired a different Seg-10 by reassortment. Although Seg-7 from NET2008/05 was also most closely related to the BTV-6 vaccine (99.7/100% nt/aa identity), the Seg-7 sequence derived from the blood sample of the same animal (NET2008/06) was identical to that of the Netherlands BTV-8 (NET2006/04 and NET2007/01). This indicates that the blood contained two different Seg-7 sequences, one of which (from the BTV-6 vaccine) was selected during virus isolation in cell-culture. The predominance of the BTV-8 Seg-7 in the blood sample suggests that the virus was in the process of reassorting with the northern field strain of BTV-8. Two genome segments of the virus showed significant differences from the BTV-6 vaccine, indicating that they had been acquired by reassortment event with BTV-8, and another unknown parental-strain. However, the route by which BTV-6 and BTV-8 entered northern Europe was not established.
Introduction
Bluetongue virus (BTV) is the ‘type’ species of the genus Orbivirus, within the family Reoviridae [1]. BTV is transmitted primarily by biting midges (Culicoides spp.) in which it also replicates, and is capable of infecting a wide range of ruminant, free ranging and captive species of wild ruminants [2], [3] as well as camelid species [4]–[7]. Some BTV strains have also been shown to be transmitted vertically, or via an oral route between individual mammalian hosts in the field [8]–[13], and after experimental infection [14], [15]. There are also recorded cases of large carnivores becoming infected after injection of BTV infected vaccines, or ingestion of infected meat [16]–[18].
Bluetongue (BT) is listed as a ‘notifiable disease’ by the Office International des Epizooties (OIE) [19], causing severe clinical signs that can include fever, lameness (coronitis), swelling of the head (particularly the lips and tongue) and death. The more severe forms of the disease are most frequently seen in sheep (particularly in the ‘improved’ mutton and wool breeds that are common in Europe) and in white-tailed deer (particularly in North America) [20]–[26]. Severe clinical signs and fatalities can occasionally also occur in cattle, goats and camelids [5], [27]–[29]. Although the infection is often inapparent in these other species, they can act as silent reservoirs, remaining viraemic for several months (particularly cattle) [25], [30]. However, even subclinical infection can carry significant costs, including loss of condition, reduced milk yield, infertility and abortion [31], as well as indirect costs associated with the export restrictions and the surveillance requirements imposed to limit the spread of the virus [32], [33].
Prior to 1998, occasional BT outbreaks had occurred in Europe, although in most cases these were relatively short lived (∼4–5 years) and involved a single BTV strain/serotype on each occasion (reviewed by Mellor et al. [34]). However, in 1998, a major series of BT incursions began in Europe, with new introductions in almost every subsequent year (to date), involving BTV-1, -2, -4, -6, -8, -9, -11 and -16 [35-38-www.reoviridae.org/dsRNA_virus_proteins/outbreaks.htm].
Live- BTV vaccines have been widely used to control the disease in the USA and as multivalent preparations (containing multiple serotypes) in Israel and southern Africa [39]–[41]. Monovalent ‘live-vaccines’ of BTV-2, 4, 8, 9 (western group) and BTV-16 (eastern group) have also been used, in attempts to minimise virus circulation in the Mediterranean region. Vector-borne spread of vaccine viruses has been reported before [42]–[44]. The release of these vaccine strains, some of which (including BTV-2 and 16) have persisted in the field, has further increased the level of genetic diversity within the European BTV population [45]–[47].
BTV incursions (from the east - possibly via Turkey and from North Africa) were initially restricted to Mediterranean Europe. However, during August 2006 sheep infected with a sub-Saharan African lineage of BTV-8, were identified in the Maastricht region of the Netherlands, the first time BTV had been detected in northern Europe [35]. This represented the start of the largest single outbreak of BT on record, subsequently spreading across the whole of Europe [35], [48], [49].
The BTV genome consists of 10 linear double-stranded RNA segments, which encode a total of 7 structural proteins (VP1 to VP7) and 3 distinct nonstructural proteins (NS1, NS2 and NS3/NS3a) [50]–[55]. The two inner layers of the BTV capsid (identified as the ‘sub-core’ and ‘core’) are composed of major structural proteins VP3 and VP7 (encoded by genome segments [Seg-] 3 and 7 respectively). The innermost subcore-shell surrounds the ten segments of the virus genome (one copy of each segment per particle) as well as three minor enzyme proteins VP1, VP4 and VP6 (encoded by Seg-1, 4 and 9 respectively). These core proteins and two of the non-structural proteins that are also synthesised in infected cells (NS1 and NS2 - encoded by Seg-5 and 8 respectively) are highly conserved and are antigenically cross-reactive between different strains of BTV. Although, the genome segments encoding these conserved proteins do show sequence-variations that reflect the geographic origin of the virus isolate (topotype), they show no significant correlation with the virus serotype [35], [53], [56]–[59]. Most BTV isolates that have been analysed can be divided into two major ‘eastern’ or ‘western’ topotypes, then into a number of further geographic subgroups, based on phylogenetic analysis of their nucleotide sequences [35], [56], [57], [60], [61].
Prior to 2008, 24 distinct serotypes of BTV had been identified, which can be distinguished in ‘neutralisation’ assays by the specificity of their reactions with the neutralising antibodies that are generated during infection of mammalian hosts. These neutralising antibodies interact with the more variable outer capsid proteins VP2 and VP5 (particularly VP2) encoded by Seg-2 and 6, respectively [1], [62]–[64]. Consequently Seg-2 and 6 vary in a manner that correlates with both the geographic origins (topotype) of the virus strain and virus serotype (particularly Seg-2), making them the most variable components of the BTV genome [35]. Different BTV ‘types’ can also be identified by conventional or real-time RT-PCR assays and/or sequence analyses targeting Seg-2 [35], [57]. Maan et al [57] showed that the Seg-2 sequencles of different BTV strains not only form distinct clades for each serotype, but also that the sequences of certain serotypes cluster more closely together, identifying ten distinct nucleotypes (identified as A–J).
In 2008 a virus was detected in goats from Switzerland (Toggenburg orbivirus - TOV), which shows a significant level of divergence in each of its genome segments, from the ‘major’ eastern and western topotypes of BTV. This virus has been provisionally identified (by serological and nucleic acid based analyses) as a novel 25th BTV serotype (BTV-25) and represents a further nucleotype (K) [[65]–see discussion].
BTV Seg-7 and 10, which encode outer-core protein VP7 and NS3/NS3a respectively, show intermediate levels of nucleotide variation between different isolates, dividing them into a number of different clades that show only partial correlation with the geographic origin of the virus [35]. VP7 can mediate surface attachment, penetration and infection of insect cells by BTV cores [66], while NS3 has been associated with the release of virus particles from insect cells [67], [68], suggesting that these proteins may collectively influence the efficiency of BTV infection and dissemination within the vector insect. It has therefore also been suggested that the variations observed in Seg-7 (VP7) and Seg-10 (NS3) may relate to transmission of the virus by distinct insect vector species/populations in different geographical regions [35], [60], [69]–[71].
By 2008 western strains of BTV-1 and BTV-8 (originating in sub-Saharan Africa) had both become established in northern Europe [34], [72], [73]. However, despite vaccination campaigns, bluetongue infected animals were again identified during September 2008, on 4 farms in the east of the Netherlands (Heeten, Luttenberg, Barchem and Oldenzaal–Figure 1) and on November 5th 2008 in Germany [74]. The virus was subsequently isolated from a bovine blood sample, taken from an animal showing clinical disease (Heeten), and was identified as BTV-6 (reported here). As part of attempts to clarify its origins, the complete nucleotide sequence of the virus genome was determined and compared to other European field strains, BTV vaccine strains used in the region, representative ‘eastern’ and ‘western’ BTV strains from other parts of the world, as well as the BTV-6 reference, vaccine and field strains from South Africa and USA.
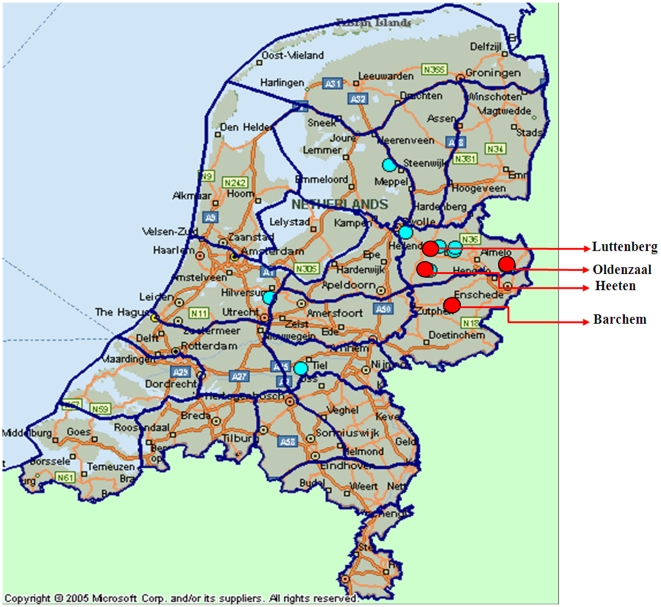
Figure 1. Map of geographical location of the BTV-6 affected farms in east of the Netherlands.
The positions of the four farms originally sampled (Heeten, Barchem, Luttenberg and Oldenzaal) are indicated by red dots. Other farms where BTV-6 was detected are indicated by blue dots.
Methods
Virus isolation and propagation in cell culture
EDTA treated blood samples from three cows (from farms at Barchem (10th October 2008), Luttenberg (16th October 2008) and Heeten (17th October 2008) in eastern Netherlands), and one serum sample (Heeten), were sent to the Community Reference Laboratory (CRL) and Arbovirus Molecular Research Group (AMRG) at the Institute for Animal Health (IAH) Pirbright (21st October 2008). These blood samples were taken from naturally infected animals in the field, by qualified veterinarians, as part of normal veterinary care and diagnostic testing procedures in the Netherlands. A sample of the blood from Heeten (stars number A163/08-3) is stored as sample ‘NET2008/06’ in the ‘dsRNA virus reference collection’ (dsRNA-VRC) at IAH Pirbright. dsRNA-VRC numbers for blood samples, or virus isolates use a generic format: ‘three letter country code, year of sampling/isolate number for that year’.
A 3.0 ml aliquot of the blood (NET2008/06) was washed three times with 10 ml of sterile phosphate-buffered saline (PBS). After each wash the red blood cells (RBC) were centrifuged at 3000×g for 5 min at 4°C and the supernatant was discarded. After the final wash, the RBCs were resuspended in 3.0 ml of PBS and RNA was extracted from 2.0 ml for RT-PCR and sequence analysis. One ml of the washed RBC was also used for virus isolation in C. sonorensis (KC) cells. At 7 dpi KC culture supernatant (KC1 - NET2008/04) was harvested and used to infect BHK-21 cells. The resulting virus isolate (KC1/BHK1 - NET2008/05) along with NET2008/04 and NET2008/06 were used for sequencing studies. Further details concerning the origins and passage history of individual virus isolates can be found at www.reoviridae.org/dsRNA_virus_proteins/ReoID/btv-6.htm.
Serum neutralisation tests
Antiserum from the Heeten farm (sample number A163/08-2) from the same cow as virus isolate NET2008/05, was also sent to CRL at IAH Pirbright (21st October 2008), for neutralising antibody testing against the 24 BTV serotypes in ‘serum neutralisation tests’ (SNT).
A standard ‘constant virus - varying serum’ method was used, with appropriate controls [75]. Briefly the test serum sample was heat inactivated at 56°C for ∼30 minutes, then serially diluted (1∶10). A fixed amount (100 VERO cell-TCID50/100 µl) of each of the 24 BTV serotypes was added to an equal volume of each serum dilution, in a 96 well plate (in four repeats), then incubated for ∼1 hour at 37°C followed by an overnight incubation at +4°C. This will allow antibodies to neutralise homologous virus. Fifty μl of a VERO cell suspension (2×105 viable cells/ml), was then added to all wells, and the plate sealed with sterile cover then placed at ∼37°C (with 5% CO2). Plates were examined under a light microscope for cytopathic effects (CPE) at 3–4 days and 6–7 days post addition of cells. The neutralising antibody level was expressed as a ‘titre’ for each serotype, representing the inverse of the final dilution of serum at which each virus was neutralised, as calculated from the duplicate assays using the Karber formula [76]. Serum neutralising antibody titres of >10 were considered significant.
Identification of BTV and molecular typing
Viral RNA for conventional and real time RT-PCR assays were extracted from cell-free infected tissue culture supernatants, and from EDTA treated blood, using the QIAamp Viral RNA Mini Kit (QIAGEN) as per manufacturer's protocol. RNA for synthesis of full-length cDNA copies of BTV genome segments was also purified from infected KC and from BHK cells using TRIzol reagent (Invitrogen) [57], [77], [78].
Serogroup-specific real-time RT-PCR assays targeting Seg-1 were carried out as described by Shaw et al. [79]. Conventional ‘type-specific’ RT-PCR assays (targeting Seg-2 of all 25 established BTV serotypes) were performed as described by Mertens et al. ([80] and–manuscript in preparation). Type specific real-time RT-PCR assays were carried out for the European BTV types, using assays kits supplied by (LSI–Laboratoire Service International), according to the manufacturer's instructions.
Terminal and internal primers that are specific for each genome segment were also used in conventional RT-PCRs, to amplify cDNAs for sequencing studies. These included type-specific and ‘nucleotype C’ specific primers targeting Seg-2 of BTV-6 (Table 1). In brief, a single-tube reaction contained the SuperScript™ III one-step RT-PCR system (Invitrogen) and high fidelity platinum® Taq was used with RNA templates extracted from blood or cell culture supernatant. The primer-template mix was heated to 95°C for 3 min to denature the viral dsRNA, followed by immediate cooling on ice, and addition of the reaction mix. Amplification of Seg-2, was carried out in 50 µL reaction volumes containing 25 µL of 2X reaction mix, 0.2 µM of each primer (1 µL of 10 µM,), 6 µL (1 pg–1 µg) of denatured RNA, 1 µL of SuperScript™ III RT/Platinum® Taq High Fidelity Enzyme Mix and nuclease free water to 50 µL. The RNA was reverse-transcribed at 55°C for 30 min. This was followed by an ‘inactivation/activation’ step at 94°C for 2 min (in order to simultaneously inactivate reverse transcriptase and activate the DNA polymerases), 40 amplification cycles (94°C for 15 sec, 55°C for 30 sec and 68°C for 1 min/kb), and a terminal extension step at 68°C for 5 min. The RT-PCR products were analysed and purified by 1% agarose gel electrophoresis (AGE) in tris acetate EDTA (TAE) buffer. The cDNA bands were stained with ethidium bromide, visualized under UV light, then excised and recovered from the gel for sequencing. Full length cDNA copies of individual BTV genome segments were also synthesised and amplified by RT-PCR, using the ‘anchor spacer–ligation’ method described by Maan et al. [35], [78] and Potgieter et al. [81].
Table 1. Primers for specific amplification of Seg-2 of BTV-6, by RT-PCR.
| Primer pairs | Individual forward and reverse primers* | Primer Sequence (5′ to 3′) | Position on genome segment 2 (nt) | Predicted Product Size (bp) |
| Primer pair I | BTV-6/2/301F | GGTGGTATGTATAGAGGAAG | 875-894 | 1631** |
| BTV-6/2/790R | ACCACGCTACTCTGTATGCC | 2506–2487 | ||
| Primer pair II | BTV-6/2/153F | CGAGGCGATTGGTGACACAGGT | 461-482 | 2226 |
| BTV-6/2/853R | CAAAGGGAACCTCGCGCGTAATC | 2687–2664 | ||
| Primer pair III | BTV-6/2/35F | GAGCGAAGATGATGAGGT | 107–124 | 1211 |
| BTV-6/2/439R | GTCTTCTTCGTCTGTGAGATCAA | 1318–1295 |
*Individual primers are identified by the BTV serotype (e.g. BTV-6) followed by the number 2 (to indicate Seg-2), then a number to indicate the relative amino acid position of the primer within VP2, followed by F or R to indicate forward or reverse orientation.
**This primer pair also amplifies Seg-2 of BTV-14 and BTV-21, the most closely related serotypes to BTV-6 within nucleotype ‘C’ and it is therefore regarded as nucleotype ‘C’ specific.
The different cDNA amplicons were purified by AGE, recovered using a GFX™PCR DNA and gel band purification kit' (Amersham Pharmacia Biotech, Inc), then sequenced using an Applied Biosystems BigDye ddNTP capillary sequencer. Consensus sequences from each segment were assembled and analyzed using SeqMan Software (DNAStar Inc.).
The sequences generated were aligned with data for BTV genome segments from GenBank (Table S1) [35], [57], [72], using CLUSTAL W software [82]. Phylogenetic trees were constructed by neighbour-joining using distance matrices, generated by the p-distance determination algorithm in MEGA version 4.1 software (500 bootstrap replicates) [83]. Sequence relatedness is reported as percentage identity. The sequences obtained for each genome segment of NET2008/05 have been submitted to GenBank (accession numbers are shown in Table 2 and 3). Additional BTV-6 and BTV-8 field, reference and vaccine strains were also sequenced and submitted to Genbank (Table 3).
Table 2. Characteristics of dsRNA genome segments (cDNA copy) and proteins of the bluetongue virus serotype 6 Netherlands (NET2008/05).
| †Genome segment (Size: bp) | ORFs (bp - including stop codon) | Protein nomenclature (§: protein structure/function) | Number of Amino acids (Da) | Location | Accession numbers | 5′ Terminal sequences of the positive strand | 3′ Terminal sequences of the positive strand |
| 1 (39 44) | 12-3920 | VP1 (Pol) | 1302 (149,476) | within the sub-core at the 5 fold axis | GQ506472 | 5′ - GTTAAA ATGCAATGGTCGCA | TGA GAGCACGCGCCGCATTACACTTAC - 3′ |
| 2 (2922) | 17-2885 | VP2 | 955 (110,315) | outer capsid | GQ506473 | 5′- GTTAAATTAGTTTCGTGATG | TAA CCTCAGTGACATGGGGTTCACGAACTAATCAACTTAC -3′ |
| 3 (2772) | 17-2723 | VP3 (T2) | 901 (103,307) | sub-core capsid layer (T = 2 symmetry) | GQ506474 | 5′- GTTAAATTTCCGTAGCCATG | TAG ATGTGCGACCAATCTATGCACTTGGTAGCGGCAGCGGGAACACACTTAC -3′ |
| 4 (1981) | 9-1943 | VP4 (Cap) | 644 (75,199) | within the sub-core at the 5 fold axis | GQ506475 | 5′- GTTAAAACATGCCTGAGCCA | TAA TGCGTGACTGCTAGGTGAGGGGGGCATGTGCAACTTAC -3′ |
| 5 (1772) | 35-1693 | NS1 (TuP) | 552 (64,445) | Cytoplasm forms tubules | GQ506476 | 5′- GTTAAAAAAGTTCTCTAGTTGGCAACCACCAAACATG | TAG TTACCGACTTCTGTTTTCTGTTTCTTCTTTTTTCTTTTTCTATTTTCTCTTAGCACTCTACTAGAACTTTTCAACTTAC -3′ |
| 6 (1637) | 28-1608 | VP5 | 526 (59,199) | outer capsid | GQ506477 | 5′- GTTAAAAAGATCCCCATGATCGCGAAAGATG | TGACAGCGGTGATCCCGGGACTTACACTTAC-3′ |
| 7 (1156) | 18-1067 | VP7 (T13) | 349 (38,587) | Outer core (T = 13 symmetry) | GQ506478 | 5′- GTTAAAAATCTCTAGAGATG | TAGTCCACTTTGCACGGGTGTGGGTTATGCGGGTGGTGTGTCGGTTGCAAGAAATATGTGTCTTGTTTAAACGTCTCTGGGATTACACTTAC-3′ |
| 8 (1125) | 20-1084 | NS2 (ViP) | 354 (40,698) | Cytoplasm, viral inclusion bodies (VIB) | GQ506479 | 5′- GTTAAAAAATCCTTGAGTCATG | TAGGCGCTTGTGACCGCGTGGTTAGGGGGGGATTTTACACTTAC-3′ |
| 9 (1049) | 16-1005 | VP6 (Hel) | 329 (35,596) | Within the sub-core at the 5 fold axis | GQ506480 | 5′- GTTAAAAAATCGCATATGTC | TAAAGGGTCCAGGGTACCTTCTTGACGTAGGGCGATTTTACACTTAC-3′ |
| 10 (822) | 20-709 | NS3 | 229 (25,556) | Cell Membrane | GQ506481 | 5′- GTTAAAAAGTGTCGCTGCCATG | TGAGGACAGTAGGTAGAGTGGCGCCCCGAGGTTTGCGTCGTGCAGGGTGGTTGACCTCGCGGCGTAGACTCCCACTGCTGTATAACGGGGGAGGGTGCGCGACACTACACACTTAC-3′ |
| 10 (822) | 59-709 | NS3a | 216 (23,966) | Cell Membrane | GQ506481 | 5′ - GTTAAA AAGTGTCGCTGCCATGCTATCCGGGCTGATCCAAAGGTTCGAAGAAGAAAGGATG | TGAGGACAGTAGGTAGAGTGGCGCCCCGAGGTTTACGTCGTGCAGGGTGGTTGACCTCGCGGCGTAGACTCCCACTGCTGTATAACGGGGGAGGGTGCGCGACACTACACACTTAC-3′ |
Pol = RNA polymerase; Cap = capping enzyme (guanylyltransferase); Hel = helicase enzyme [42]; T2 = protein with T = 2 symmetry; T13 = Protein with T = 13 symmetry; ViP = viral inclusion body matrix protein; TuP = tubule protein. Letters in bold blue are the 5′ and 3′ terminal conserved sequences in genome segments of BTV-6 NET2008/05.
Table 3. Accession numbers of BTV strains sequenced and used for phylogenetic analyses.
| IAH Reference collection number or Year of isolation or strain designation | Country of origin | Seg-1 | Seg-2 | Seg-3 | Seg-4 | Seg-5 (NS1) | Seg-6 (VP5) | Seg-7 | Seg-8 | Seg-9 | Seg-10 |
| BTV-6 and 8 full genomes sequenced in these studies | |||||||||||
| BTV-6 Vaccine strain | S. Africa | GQ506506 | GQ506507 | GQ506508 | GQ506509 | GQ506510 | GQ506511 | GQ506512 | GQ506513 | GQ506514 | GQ506515 |
| BTV-6 NET2008/05 | Netherlands | GQ506472 | GQ506473 | GQ506474 | GQ506475 | GQ506476 | GQ506477 | GQ506478 | GQ506479 | GQ506480 | GQ506481 |
| BTV-6 Underberg S. Africa (P635- isolated from midges) | S. Africa | GQ506488 | GQ506489 | GQ506490 | GQ506491 | GQ506492 | GQ506493 | GQ506494 | GQ506495 | GQ506496 | GQ506497 |
| BTV-6 Prototype vaccine strain (5011-60E) | S. Africa | GQ506526 | GQ506527 | GQ506528 | GQ506529 | GQ506530 | GQ506531 | GQ506532 | GQ506533 | GQ506534 | GQ506535 |
| BTV-6 RSArrrr/06 | S. Africa | GQ506498 | AJ585127 | GQ506499 | GQ506500 | GQ506501 | AJ586703 | GQ506502 | GQ506503 | GQ506504 | GQ506505 |
| BTV-6 S. Africa (BT-2-03 - isolated from a lamb) | S. Africa | GQ506516 | GQ506517 | GQ506518 | GQ506519 | GQ506520 | GQ506521 | GQ506522 | GQ506523 | GQ506524 | GQ506525 |
| BTV-6 USA2006/01 | USA | GQ506536 | GQ506537 | GQ506538 | GQ506539 | GQ506540 | GQ506541 | GQ506542 | GQ506543 | GQ506544 | GQ506545 |
| BTV-8 NET2007/01 | Netherlands | GQ506451 | GQ506452 | GQ506453 | GQ506454 | GQ506455 | GQ506456 | GQ506457 | GQ506458 | GQ506459 | GQ506460 |
| BTV-6 partial genomes sequenced in these studies | |||||||||||
| BTV-6 GER2008/05 | Germany | x | GQ506461 | x | GQ506462 | GQ506463 | GQ506464 | GQ506465 | GQ506466 | GQ506467 | x |
| BTV-6 NET2008/04 | Netherlands | GQ506468 | GU550956 | GQ506469 | GQ506470 | x | x | GQ506471 | x | x | x |
| BTV-6 NET2008/06 | Netherlands | x | GU550957 | x | x | GQ506482 | GQ506483 | GQ506484 | GQ506485 | GQ506486 | GQ506487 |
| Published partial genome used in these studies | |||||||||||
| BTV-25 (Toggenburg Orbivirus [TOV]) | Switzerland | GQ982522 | EU839840 | GQ982523 | GQ982524 | EU839841 | EU839842 | EU839843 | EU839844 | EU839845 | EU839846 |
Results
Identification of BTV in the Netherlands
Blood samples were taken during October 2008, from three cows showing mild clinical signs, consistent with BTV infection, from different farms in the eastern Netherlands (at Heeten, Luttenberg and Barchem). Two if these animals (Heeten and Barchem) had been vaccinated against BTV-8 during May/June 2008. A fourth apparently healthy cow was also sampled as part of routine pre-export testing from a farm at Oldenzaal. These samples tested positive for the presence of BTV RNA by RT-PCR (targeting Seg-10). Partial sequence analyses of Seg-10 showed that although one of the samples (Luttenberg) contained a “mixed sequence” (and was therefore unreadable), the other three samples (from Heeten, Barchem and Oldenzaal) showed significant sequence differences when compared to the previous northern European outbreak strain of BTV-8 (∼95% identity over a 200 base pair region [35]) and BTV-1 (82.0–83.3% identity over full length region). These PCR results were confirmed by re-sampling and re-testing of the RT-PCR positive animals.
Virus isolation and propagation in cell culture
A sample of blood taken from a cow in eastern Netherlands (Heeten) (sample number A163/08-3) in October 2008, which had tested positive for bluetongue in a Seg-10 based real time RT-PCR assay conducted at the Central Veterinary Institute of Wageningen UR (CVI), Lelystad, was sent to the CRL at IAH Pirbright in October 2008 (stored as dsRNA-VRC sample number NET2008/06). The virus was isolated (single passage) in KC cells without CPE (generating isolate NET2008/04) then passaged in BHK-21 cells (isolate NET2008/05), causing 100% CPE at 4 days post infection. Subsequent real-time RT-PCR assays (at IAH) targeting BTV Seg-1 [79], gave CT values of 26 for the Heeten blood sample (NET2008/06), and 12–35 for cell-culture supernatants from isolates NET2008/05 and NET2008/04, confirming the presence of BTV.
Identification and typing of BTV-6
Conventional RT-PCR assays (at CVI) using primers directed against Seg-2 of the BTV serotypes recently detected in Europe (BTV-1, 2, 4, 8, 9 and 16 [80] see: www.reoviridae.org/dsRNA_virus_proteins/ReoID/BTV-S2-Primers-Eurotypes.htm) failed to amplify any cDNA products of the appropriate sizes from the four bovine blood samples previously identified as positive by BTV group-specific RT-PCR. This indicated that the new virus represented an additional European serotype. However, subsequent RT-PCR assays (at IAH) using two sets of experimental primers targeting Seg-2 from each of the BTV serotypes (1 to 25) (Mertens et al., [80] and manuscript in preparation), generated cDNA amplicons of the expected sizes from two of the three blood samples (Heeten and Barchem), but only with the BTV-6 specific primers. These analyses not only identified the virus as BTV-6, but also excluded the other 24 BTV serotypes, providing the first positive identification of this type anywhere in Europe. However, one of the blood samples (Luttenberg) from an unvaccinated cow gave positive results with both the BTV-6 and BTV-8 specific primers, indicating that this animal had recently been infected with both serotypes.
RT-PCRs using two further pairs of BTV-6 specific primers and one pair of primers specific for ‘nucleotype C’ (which includes BTV-6, 14 and 21) (Table 1) were used to generate cDNAs of the expected sizes from virus isolates NET2008/04 and NET2008/05 (derived from the Heeten blood sample), representing a total of ∼75% of Seg-2. This confirmed the identification of BTV-6 and provided cDNAs for sequence analyses (Figure 2). Type-specific real-time RT-PCR assays (supplied by LSI), targeting Seg-2 of BTV-1, 2, 4, 6, 8, 9, 11, 16 and 25, also subsequently generated positive results for BTV-6 from these isolates (CT value of 38 [NET2008/04] and 14 [NET2008/05]), as well as from the original Heeten blood sample (CT value of 27 [NET2008/06]).
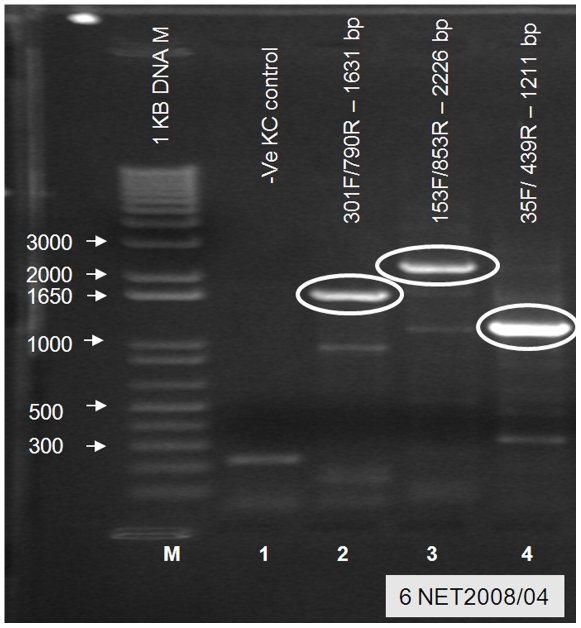
Figure 2. Electrophoretic analysis of cDNA products generated from Seg-2 of NET2008/04 using nucleotype ‘C’ and BTV-6 specific primer pairs.
PCR amplicons were generated from cDNA of BTV-6 isolate NET2008/04 with one nucleotype ‘C’ (lane 2) and two type 6 specific primer pairs (lanes 3 and 4 – Table 1). No specific amplification was seen with these primers from mock KC cells (Lane 1). Lane M: 1 kb marker.
Typing of BTV specific antibodies in sera from the Netherlands
Antiserum from the BTV-8 vaccinated cow from the Heeten farm (sample number A163/08-2), was also tested using serum neutralisation tests (SNT) against reference strains of BTV serotypes 1 to 24, and gave high neutralisation titres against both BTV-6 and BTV-8 (in each case at a neutralisation titre of 1/1280). Although, lower titres were also detected against two other serotypes (1/80 [BTV-14] to 1/120 [BTV-18]), these results are consistent with sequential infection by BTV-6 and either infection or vaccination against BTV-8 [84], [85].
Full-length sequence analyses and comparison of the NET2008/05 genome
In an attempt to identify the origins of the BTV-6 strain from Heeten, cDNAs were amplified and used to generate full length sequence data for individual genome segments of the virus (segments 1, 3, 4, 7 were sequenced from KC cell (KC1) isolate, NET2008/04; while the whole genome was sequenced from the KC1/BHK1 isolate, NET2008/05, and segments 2 (partial), 5 to 10 were sequenced from the Heeten blood sample NET2008/06). The consensus sequences obtained for most of the genome segments were identical in each case, except for Seg-7 and Seg-10 (see below). The remainder of the paper therefore refers to the sequence of the Netherlands BTV-6 KC1/BHK1 isolate (NET2008/05), except where specific changes were detected between the different samples.
Genome segment 1
Seg-1 of BTV-6 NET2008/05 was compared to other ‘eastern’ and ‘western’ BTVs, including other strains of BTV-6 sequenced in this study (Table 2 & 3). In each case Seg-1 is 3944 base pairs (bp) long, encoding the 1302 amino acid (aa) RNA dependent RNA polymerase (RdRp - Pol) (Table 2), and is highly ‘conserved’, showing >78.7% overall identity. Seg-1 of BTV-6 NET2008/05 was most closely related to the South African BTV-6 vaccine strain (5011-60E-VP1) (99.9% nucleotide [nt] identity), a BTV-6 strain from Germany (GER2008/05) (99.9% identity), and the BTV-6 reference strain (RSArrrr/06–99.8%).
The deduced aa sequence for BTV VP1 is also highly conserved, showing >93.2% identity, with a clear separation between ‘eastern’ and ‘western’ virus groups/topotypes (with a maximum of 5.8/1.7% and 12.0/3.8% nt/aa intra-topotype variation respectively within these two groups) (Figure 3) [35]. The level of nt/aa identity in Seg-1/VP1 between these major eastern and western virus groups was 78.7 to 80.7/93.2 to 95.4% respectively (Figure 3). However, BTV-25 (TOV) does not cluster with either group, showing <75.8/88.1% and <75.8/88.2% nt/aa identity with the eastern or western BTV strains respectively, suggesting that it represents a further distinct (western) group/topotype.
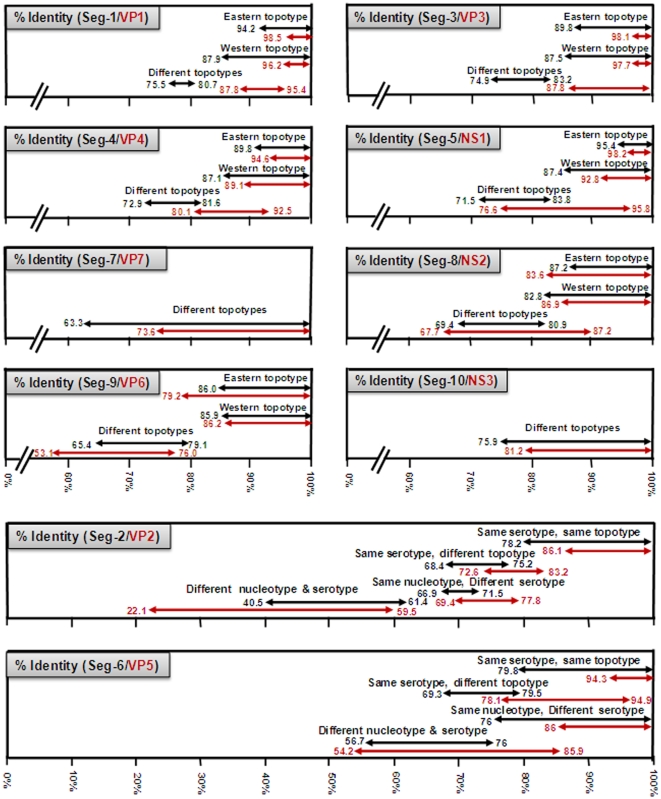
Figure 3. A graphical representation of the levels of nucleotide and amino acid sequence identities detected in different genome segments and proteins, within and between different BTV serotypes and topotypes.
Estimates of the levels of identity in each genome segment/protein, between different BTV strains are based on multiple datasets for widely distributed isolates, as described previously by Maan et al. [35], [57], [72] and listed in tables 3 and S1. The values presented here show the levels of identity detected within the ‘major’ eastern and western topotypes for each genome segment (black) and protein (red). The levels of identity that are shown between different topotypes, include data for all genome segments of BTV-25 (Toggenburg orbivirus), as well as Seg-3 of BTV-15 Australia (Ac. No. AY322427) and Seg-5 of BTV-20 Australia (Ac. No. X56735), as representatives of distinct topotypes.
Seg1/VP1 from the BTV-6 Netherland isolates was identical to that of the BTV-6 vaccine strain, with 99.8% identity to the reference strain (RSArrrr/06) from which the vaccine was originally derived [39]. Seg-1/VP1 of BTV-6 NET2008/05 was also closely related to those of other BTV-6 strains from South Africa (95.1 to 95.2% nt and 98.6 to 98.8% aa identity), but somewhat less closely related to the western strain of BTV-6 from the USA (USA2006/01–88.9/98.4% nt/aa identity).
Genome segment 2
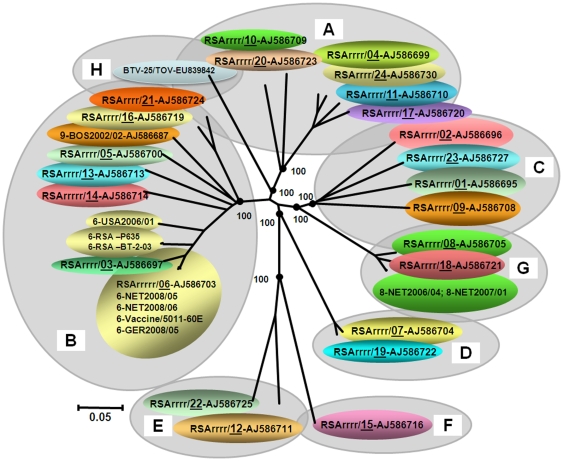
Multiple cDNAs were amplified from Seg-2 of NET2008/05 and sequenced (full–length), showing that like other BTV-6 strains, it is conserved at 2922 bp, encoding VP2, which is 955 aa long (Table 2). Comparisons with Seg-2/VP2 from the reference strains of BTV 1 to 25 (Table 3) [57], [65], [72], showed that the Netherlands strain of BTV-6 groups within ‘Seg-2 nucleotype C’ along with reference strains of BTV-6, 14 and 21 (Figure 3 and 4). It is very closely related to the reference strain of BTV-6 (99.8/99.7% nt/aa identity) and is identical to the vaccine strain of the same serotype from South Africa (Figure 4). This analysis not only confirmed its initial identification of BTV-6, but also demonstrates that NET2008/05 was derived from the ‘vaccine’ strain.
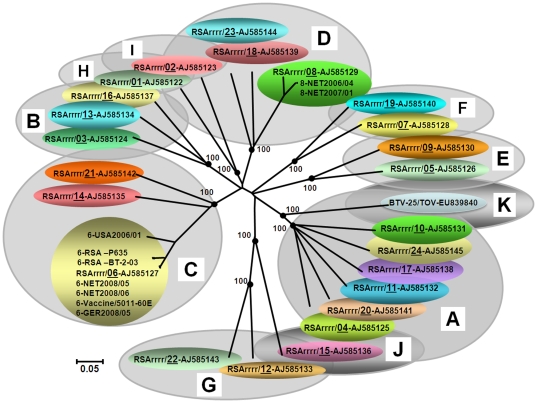
Figure 4. Neighbour-joining tree showing relationships between Seg-2 of NET2008/05 with other BTV-6 isolates and the twenty-five reference strains of different BTV serotypes.
The tree was constructed using distance matrices, generated using the p-distance determination algorithm in MEGA 4.1 (500 bootstrap replicates) [83]. The ten evolutionary branching points are indicated by black dots on the tree (along with their bootstrap values), which correlate with the eleven ‘Seg-2 nucleotypes’ designated A–K. BTV-25 (Toggenburg orbivirus [TOV]) forms a new 11th Seg-2 nucleotype (K). Members of the same Seg-2 nucleotype, are characterised by >66.9% identity in their Seg-2 nucleotide sequences, while members of different nucleotypes show <61.4% identity in Seg-2 [57], [72] (see Figure 3). The trees shown in Figures 4- 7 were drawn using same parameters.
Comparisons to sequence data for Seg-2 from multiple subsequent isolates of BTV-6 from northern Europe (2008-2009 - including GER2008/05) showed that they all group closely with NET2008/05 (>99.9/100% nt/aa identity)–indicating a common origin, with only 89.1/95% nt/aa identity to BTV-6 strain from USA (USA2006/01). Field isolates of BTV-6 from South Africa (Table 3) were also closely related to each other (with 99.3/99.5% nt/aa identity) but more distantly related to NET2008/05 (95.7 to 95.9% nt and 98.5 to 98.6% aa identity) or USA2006/01 (88.6 to 88.7% nt and 94.7 to 94.8% aa identity–Figure 4). No BTV-6 strains belonging to the major eastern topotypes/origins were available for comparison.
Genome segment 3
Seg-3 of the different European BTV strains is 2772 bp long (regardless of serotype or topotype), encoding the 901 aa of the highly conserved BTV sub-core-shell protein, VP3(T2) (Table 2). The eastern and western virus isolates formed two distinct clusters (major topotypes), which have nt/aa identities of >89.80/98.1% (eastern topotype) and 87.5/97.7% (western topotype) respectively. The overall level of nt/aa identity in Seg-3/VP3 between these eastern and western virus groups, was 79.3 to 82.4/96.9 to 99.3% respectively (Figure 3). However a single Australian strain of BTV-15 (Ac. No. AY322427) appears to represent a further distinct ‘far eastern’ group, showing <80.8/97.9% nt/aa identity with the other eastern strains and 78.9 to 82.4/95.4 to 97.6% nt/aa identity with western strains. In a similar manner, BTV-25 (TOV) shows <77/88.9% and <76.6/89.5% nt/aa identity, respectively, with the major eastern and western topotypes of BTV, again indicating (as with Seg-1/VP1) that it represents a further distinct (western) group/topotype.
Seg-3/VP3 of BTV-6 NET2008/05 showed >88.5% nt identity to the other European viruses within the major ‘western’ topotype, but only 79.8 to 83.2% identity to the ‘eastern topotype’. Within the western group Seg-3/VP3 of NET2008/05 was most closely related to the BTV-6 vaccine and reference strains (99.8/99.9% nt/aa identity), and more distantly related to South African field isolates of BTV-6 (94.6 to 94.7/99.4 to 99.6% nt/aa identity), or the North American BTV-6 (USA2006/01 - 90.1/97.2% nt/aa identity). The Netherlands virus has a unique aa change in VP3, when compared to the BTV-6 vaccine or reference strains, at position 266 (Valine to isoleucine).
Genome segment 4
Seg-4 of BTV is consistently 1981 nt in length, encoding the 644 aa of the highly conserved BTV capping and transmethylase enzyme - VP4(CaP) (Table 2). The full-length sequence of Seg-4/VP4 from BTV-6 NET2008/05 grouped with other ‘western’ BTV strains (showing >89.6/95.3% nt/aa identity), but only 79.4 to 79.9/90.4 to 91.8% nt/aa identity to the ‘eastern’ isolates.
Analysis of Seg-4/VP4 sequences again divided the majority of the isolates compared, into either an eastern or a western topotype, with nt/aa identities of >89.8/94.6% (eastern topotype) and 87.1/89.1% (western topotype) respectively. The level of nt/aa identity between these major topotypes was 77.7 to 81.6/86.1 to 92.5% (Figure 3). However, BTV-25 (TOV) showed <73.4/82.1% and <73.4/82% nt/aa identity with the eastern and western BTV strains respectively, which is consistent with its membership of a further distinct (western) group/topotype.
Seg-4/VP4 of BTV-6 NET2008/05 show 99.9/100% and 99.8/100%, nt/aa identity, to the South African vaccine and reference strains of type 6 (respectively), indicating a common origin. Like several of the other genome segments, Seg-4 of BTV-6 NET2008/05 also showed a slightly more distant relation with the BTV-6 field strains from South Africa (93.7–93.8/98% nt/aa identity), and the American BTV-6 (USA2006/01 - 90.1/97.2% nt/aa identity). Four unique aa changes were detected in the Netherlands and vaccine or reference strains of type 6, as compared to other western BTV isolates (at positions 54 (V-I), 117 (F-Y), 166 (I-T) and 825 (I-V)).
Genome segment 5
Seg-5 of BTV- 6 NET2008/05 is 1772 nt long, encoding the 552 aa of the NS1 tubule protein (TuP), with upstream and downstream NCRs of 34 and 81 bp in length (Table 2). The downstream NCR of BTV Seg-5 varies in length, even between isolates of the same serotype [35]. Seg-5 of the BTV-6 isolates from the Netherlands and South Africa has an 85 bp long 3′ NCR, while that BTV-6 from the USA (USA2006/01) is only 78 bp.
Seg-5/NS1 from the majority of the different BTV isolates compared also segregate into two ‘major’ groups, with nt/aa identities of >95.4/98.2% (eastern topotype) and >87.4/92.8% (western topotype) respectively (Figure 3). The overall level of nt/aa identity between these major topotypes was 81.6 to 83.8/92.4 to 95.8%.
However a single Australian strain of BTV-20 (Ac. No. X56735) appears to represent a further distinct eastern group, showing <81.5/92.4% nt/aa identity with the other eastern strains and 78.9 to 80.9/89.7 to 91.7% nt/aa identity with western strains. BTV-25 (TOV) shows <74.3/78.1% and <74.0/78.6% nt/aa identity with the eastern and western strains of BTV respectively, again suggesting that it represent a further distinct (western) group/topotype.
Seg-5/NS1 of BTV-6 NET2008/05 belong to the major western topotype, showing 100% identity with the South African BTV-6 vaccine and reference strains (again indicating a common origin), with 92.9/99.3% and 88.2/96.7% nt/aa identity to other BTV-6 strains from South Africa and USA respectively. Seg-5/NS1 of BTV-6 NET2008/05 also showed 98.8%, 98.6%, and 98.1% nt identity, to south African vaccine strains of BTV-1, 9 and 4 respectively, and up to 94.2/100% nt/aa identity to BTV-8 strains from Europe.
Genome segment 6
Seg-6 encodes VP5, the smaller of the two outer capsid components and second most variable of the BTV proteins. Although variations in Seg-6/VP5 can also be used to identify eight nucleotypes (A-H - Figure 5, see discussion[65], [72]), unlike Seg-2/VP2, they show only partial correlation with BTV serotype. This is demonstrated by the close relationships between BTV-7 and 19 within nucleotype D, and the more distant relationship between the reference strain of BTV-9 in nucleotype C, and the Bosnian strain of BTV-9 in nucleotype B (Figure 5). The levels of nt/aa identity in Seg-6/VP5 between different serotypes that are also present in different nucleotypes are 56.7 to 76/54.2 and 85.9% respectively. However different serotypes within the same nucleotype can show very high levels of identity approaching 100% (Figure 3). Seg-6 of BTV types 1, 2, 9 and 16 all show evidence for separation into eastern and western topotypes, [35], [72]. In contrast the BTV-4, -6 and -8 strains that have been analysed are exclusively from western origins, and consequently there is (as yet) no evidence for separation of their Seg-6 sequences into eastern and western topotypes.
Figure 5. Neighbour-joining tree showing relationships between Seg-6 of NET2008/05 with other BTV-6 isolates and the twenty-five reference strains of different BTV serotypes.
The seven evolutionary branching points are indicated by black dots on the tree (along with their bootstrap values), dividing the sequences into eight ‘Seg-6 nucleotypes’ designated ‘A–H’. In previous studies [72], six Seg-6 nucleotypes were identified, however the additional analyses described here indicate that the previous ‘nucleotype C’ should be subdivided (into nucleotypes ‘C’ and ‘G’). BTV-25 (Toggenburg orbivirus [TOV]) forms a new 8th Seg-6 nucleotype (H). Members of the same nucleotype show >76% nt identity in Seg-6, while members of different nucleotypes show <76% nt identity (see Figure 3).
Seg-6/VP5 of the BTV-6 isolates analysed (Table 2 and 3), are conserved at 1637 bp/526 aa long, with >89.3/98.5% nt/aa identity overall. Seg-6 of NET2008/05 grouping within ‘nucleotype B’, with reference strain of BTV-3 from South Africa (RSArrrr/03–nt/aa identity of 96.1 to 96.5/99.6%) and BTV-6 from the USA (USA2006/01–nt/aa identity of 88.9 to 89.3/98.7%) (Figure 5). The majority of these BTV-6 isolates (with the exception of BTV-6 USA2006/01) belong to an African sub-group, with >94.6/99.4% nt/aa identity. However, the European strains (represented by NET2008/05) were even more closely related to the reference strain of BTV-6 (RSArrrr/06) with only 5 nt and 2 aa differences (99.7/99.6% nt/aa identity) and are identical to the BTV-6 vaccine strain (RSAvvvv/06). This confirms the initial identification of the northern European isolate as type 6 that was made by Seg-2 RT-PCR. Subsequent analyses of multiple northern European isolates of BTV-6 (including GER2008/05) show that they all contain Seg-6 that is identical to that of NET2008/05, and therefore all appear likely to be derived from the same origin.
Recent South African isolates of BTV-6 (Table 3) are closely related to each other, with >99.7/100% nt/aa identity, but are more distantly related to NET2008/05 (with only 94.6 to 94.8/99.4% nt/aa identity), or BTV-6 from USA (USA2006/01) (89.5 to 89.6/98.5% nt/aa identity) (Figure 5), indicating that they do not share a very recent common ancestry with the northern European isolate.
Genome segment 7
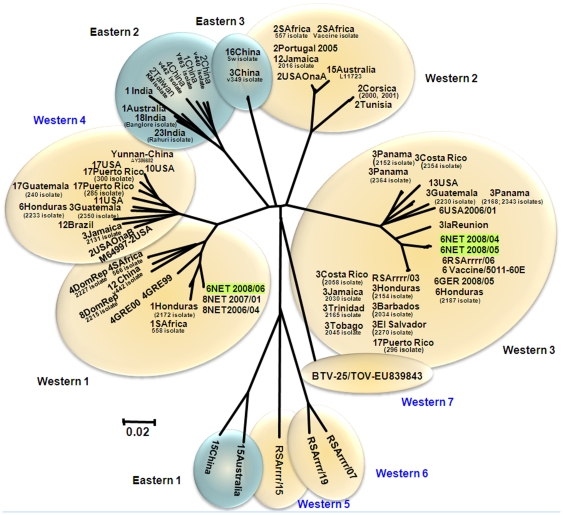
Seg-7 of NET2008/05 is 1156 bp long, encoding 349 aa of the major BTV serogroup-specific antigen and core surface protein - VP7 (Table 2). The aa sequence of VP7 is significantly more conserved (>73.6% identity) than the nt sequence (>63.3%) reflecting large numbers of synonymous mutations in the third base position (Figure 3) [35]. Seg-7 was reported to form six distinct clusters: three of these are primarily from western origins (western 1, 2 and 3) and three from an eastern origin (eastern 1, 2 and 3) [35]. However, Seg-7 from the Chinese strain of BTV-12 groups within western group 1, suggesting some movement of strains between geographic regions. Analysis of additional isolates from around the world also identified four additional western clusters, as well as an isolate from Yunnan, China (AY386682) in western group 4, and BTV-15 from Australia (Ac. No. L11723) within western group 2 (Figure 6). BTV-25 (TOV), showed 70 to 79.1%/79.9 to 93.4% nt/aa identity with other BTV strains, representing a further distinct group/topotype and forms a western group 7.
Figure 6. Neighbour-joining tree showing relationships between Seg-7 of NET2008/05 with other BTV-6 isolates and other BTV strains from different serotypes from around the world.
The nt/aa sequence of Seg-7/VP7 of NET2008/04 (KC isolate) and NET2008/05 (KC1/BHK1 isolate) showed >70.5/99.7% nt/aa identity to other BTV strains, and as observed with the majority of other segments (except Seg-10), highest levels of identity to the BTV-6 vaccine strains in ‘western cluster 3’. However, in contrast Seg-7 from NET2008/06 showed 79.3 to 79.9/94.5 to 94.8% nt/aa identity with other BTV-6 strains, but 100% nt/aa identity with BTV-8 from the Netherlands 2006/2007 (NET2006/04 and NET2007/01), within ‘western group 1’ (Figure 6).
Genome segment 8
Seg-8 of BTV is conserved at 1125 bp, encoding the 354 aa of the viral inclusion body (VIB) matrix protein - NS2 (Table 2). Seg-8 of the BTV isolates also showed clear separation into major eastern and western topotypes, with NET2008/05 grouping along with other western viruses, including the vaccine strains of BTV-2, 4 and 9 and European field strains of BTV-2 and 4 (data not shown). As observed with the other conserved segments, BTV-25 (TOV) forms a second distinct western topotype.
The BTV isolates that were compared from eastern origins, all showed nt/aa identities in Seg-8/NS2, of >87.2/83.6%, while the ‘western’ viruses showed >82.8/86.9% identity (Figure 3). The only exception was BTV-25 (TOV), which although showed <70.4/70.1% nt/aa identity with eastern isolates, only <71.1/69.5% nt/aa identity with the other western BTV strains. Overall, nt/aa identity levels of 75.0 to 80.5/80.0 to 87.2% were detected between the ‘major’ eastern and western topotypes of BTV (Figure 3). The results obtained, like those for the other conserved genome segments are consistent with BTV-25 (TOV), as a member of a further distinct (western) topotype (Figure 3).
Seg-8 of NET2008/05 and NET2008/06 showed >99.9% nt and aa identity to the vaccine strains of BTV-6 and to other BTV-6 isolates from Germany (GER2008/05). Although clearly distinct, the Netherlands strain also showed relatively high levels of nt/aa identity to South African field strains of BTV-6 (96/98%), and to BTV-6 from USA (USA2006/01–88.8/93.7%).
Genome segment 9
Seg-9 codes for VP6, a minor core protein and the helicase enzyme (Hel) of BTV. As in previous studies [35], the BTV Seg-9 nt sequences analysed here were clearly divided into eastern and western topotypes. Seg-9 of the eastern BTV isolates is 1052 bp, encoding a protein 330 aa in length, while Seg-9 from the western lineage is 1049 bp (329 aa) (Table 2) [35]. Of the 112 BTV strains analysed, 96 isolates had two in frame initiation codons, at nucleotides 16 to 18 and 28 to 30 of Seg-9, which can generate two related forms of VP6. This agrees with previous observations of two closely migrating VP6 bands in purified BTV particles or in in vitro translation studies [86]. Only 16 isolates (belonging to both eastern and western topotypes) had a single start codon at nucleotides 28 to 30.
The level of nt/aa identity in Seg-9/VP6 within the ‘major’ eastern and western topotypes was >86.0/79.2% (eastern topotype) and >85.9/86.2% (western topotype) respectively, with 69.1 to 79.1/57.3 to 76.0% nt/aa identities between these groups (Figure 3). However, BTV-25 (TOV) showed <71.4/66.2% nt/aa identity with other western isolates; <71.8/66.6% with most eastern BTV isolates, but only (65.4/53.1% nt/aa) to BTV-15 Australia (Ac. No. DQ289044). As with the other genome segments analysed, these data suggest that BTV-15 Australia and BTV-25 (TOV) represent further distinct ‘far eastern’ and western BTV topotypes respectively.
Seg-9 of NET2008/05 belongs to the major western topotype and is identical to that of the South African BTV-6 vaccine, but shows minor differences (99.7/99.4% nt/aa identity) when compared to Seg-9/VP6 of the reference strain, from which the vaccine strain was derived. It is also closely related to but distinct from BTV-6 strains from South Africa (94.4 to 95.3/93.6 to 94.8% nt/aa identity), and the USA (USA2006/01 - 90.0/91.0% nt/aa identity).
Genome segment 10
Seg-10 of NET2008/05 is 822 bp long and codes for two small, related non-structural proteins, NS3 (229 aa) and NS3a (216 aa) (Table 2). Seg-10/NS3 sequences can be divided into five groups, with >75.9/81.2% nt/aa conservation overall (Figure 3 and 7). These include three major groups, two from western and one from eastern origins containing the majority of Seg-10 sequences, as well as minor eastern and western groups each containing sequences from only a single isolate (type 15 from China and BTV-25 [TOV] respectively).
Figure 7. Neighbour-joining tree showing relationships between Seg-10 from BTV-6 NET2008/05 with multiple other BTV-6 isolates and other BTV strains of different serotypes from around the world.
The majority of the western viruses (western groups 1 and 2) showed 80.0 to 97.4%/90.4 to 99.6% nt/aa identity to eastern viruses (eastern groups 1 and 2). However ‘western group 3’ represented by the newly discovered BTV-25 (TOV), showed a maximum of 79.1 to 79.3 nt and 83.4% aa identity with the eastern and western isolates respectively (Figure 3 and 7).
Seg-10 from European isolates of BTV-1, 9 and 16 cluster together in eastern group 1, while European isolates of BTV-1, 2 and 4, and South African reference strain of type 1 cluster within western group 2 along with BTV-8 reference strain, and vaccine strains of BTV-8, 11 and 17. Seg-10 of the South African vaccine strains of BTV-1, 2, 3, 4 and 6, cluster within western group 1 along with BTV-8 NET2006/04 and BTV-2 and 6 reference strains and field strains of BTV-2, 6 and 12 from Portugal, USA and Jamaica. NET2008/05 is also included in this western group 1 and was most closely related (98.4/99.6% nt/aa identity) to the reference (Ac. No. AF481094) and vaccine strains of BTV-2 (Ac. No. AF481094), a field strain of BTV-2 from Portugal (EF434179/PT/26629/05) and another field strain of BTV-12 from Jamaica (Ac. No. AY426595). Seg-10 of NET2008/05 and NET2008/06 was more distantly related (93.6/99.6% nt/aa identity) to the BTV-6 vaccine/reference strain and BTV-6 strain from USA (USA2006/01–93.0/99.6%) in western group 1.
Discussion
In September 2008, cows on three different farms in the eastern Netherlands two of which had previously received an inactivated BTV-8 vaccine, displayed mild clinical signs of BT (particularly coronitis). A fourth apparently healthy cow that was tested for BTV prior to international transport was also RT-PCR positive. Blood samples from these animals gave positive results for the presence of BTV RNA using RT-PCR targeting Seg-10 (at CVI). Partial sequence-analyses of Seg-10 from these four samples showed significant differences when compared to strains of BTV-1 (17.3%) and BTV-8 (3.3%) from northern Europe. These initial data indicated that the virus represented a new introduction to the region.
Conventional RT-PCR assays, using primers directed against Seg-2 of BTV-1, 2, 4, 8, 9 and 16 failed to amplify any products of an appropriate size, providing the first indication that the virus also represented a serotype not previously detected in Europe. Further RT-PCR assays using primers for the 25 established BTV types (1 to 25) identified the virus as BTV-6, although one of the original blood samples from an unvaccinated animal (Luttenberg) also gave positive results with primers targeting Seg-2 of BTV-8, indicating a mixed infection. This demonstrates that both BTV-6 and BTV-8 were circulating in the region. Real-time RT-PCR assays targeting Seg-2 of BTV-1, 2, 4, 6, 8, 9, 11, 16 and 25 (supplied by LSI) subsequently also identified BTV-6 (CT values 14, 27, and 38) in both the blood sample and the virus isolates from Heeten, demonstrating that this strain of BTV-6 could be detected and identified by these assays.
Neutralisation assays demonstrated the presence of antibodies to both BTV-6 and 8 in serum from the infected animal at Heeten, reflecting infection with BTV-6 as well as infection and/or vaccination with BTV-8.
BTV-6 had previously been isolated from Africa, Middle East, India, Pakistan, USA, Central and South America [87] and therefore exists as both eastern and western strains, although no sequence data for the eastern strains were available for these analyses. The data presented here represent the first complete sequence (segments 1-10) of the northern European strain of BTV-6, as well as BTV-6 field and vaccine strains from South Africa and a BTV-6 field strain from North America (USA2006/01).
Sequence analyses of Seg-2 from the Heeten blood sample (NET2008/06) and isolate (NET2006/05) showed that it clusters in ‘Seg-2 nucleotype C’ (identified by Maan et al. [57], [72]–Figure 4), and revealed 5 nt differences (99.8/99.7% nt/aa identity) from the reference strain of BTV-6 (RSArrrr/06 - isolated in South Africa in 1958: reviewed by Alpar et al. [39]). This not only confirmed the virus type, but also indicated that it belongs to the major ‘western’ topotype, and groups with other sub-Saharan African strains of BTV-6. Further comparisons of Seg-2/VP2, showed that it is identical to the South African live attenuated vaccine strain of BTV-6, providing the first indication that it was in fact derived from this source.
Seg-2 and Seg-6 of TOV [65] were also included in these analyses, showing levels of variation that are consistent with its inclusion as BTV-25, as a representative of a distinct eleventh nucleotype of Seg-2 (nucleotype K - Figure 4) and an eighth nucleotype of Seg-6 (nucleotype H–Figure 5). Comparisons of VP3 from the Toggenburg orbivirus isolate [65] showed 88.0 to 89.5% aa identity with other BTV strains. This is significantly higher than the 77.3 to 77.9% identity which is shares with isolates of epizootic haemorrhagic disease virus (EHDV) [88], the most closely related of the other Orbivirus species to BTV [1]. BTV and EHDV themselves also show only 78.5 to 81.1% aa identity in VP3. These values compare with 91%, the lowest level of identity previously detected in VP3 within a single Orbivirus species [89]. If TOV is to be considered as a new strain/serotype of BTV, this pushes down the level of identity for VP3 within a single species, although this is perhaps inevitable as more distinct and diverse isolates are analysed and compared. Further confirmation that TOV is a member of the species Bluetongue virus could be generated by a demonstration that it can exchange/reassort genome segments with other strains of BTV [1]. However, attempts to adapt the virus to cell culture have so far been unsuccessful [65], making these experiments more difficult.
Phylogenetic analyses of the other genome segments from NET2008/05 showed that most were very similar or identical to the South African live-vaccine strain of BTV-6 (99.7–100% nt/aa identity). They also contained significantly higher numbers of nucleotide changes when compared to sequences from field strains of other serotypes already circulating in Europe, or the other live-vaccine strains that had been used in the Mediterranean region. Subsequent data for BTV-6 isolated in Germany during 2008 (GER2008/05) showed small numbers of nucleotide changes in several of genome segments, when compared to NET2008/05, but confirmed that the different isolates were all very closely related representing a single virus lineage. All of the genome segments of NET2008/05 group with those of other ‘western’ viruses, belonging to sub-Saharan African lineages. However, Seg-10 of NET2008/05 was less closely related than the other segments to that of the BTV-6 vaccine strain (93.6/99.6% nt/aa identity) and was clearly distinct from previously published data for other field strains from Europe or elsewhere. Seg-10 of NET2008/05 was most closely related to that of the South African vaccine and reference strains of BTV-2, showing 98.4/99.6% nt/aa sequence identity. This indicates that they share a recent common ancestry and suggests that the Netherlands virus acquired Seg-10 by reassortment, possibly with another BTV vaccine strain. Although it is possible that this segment was acquired during co-infection by BTV-2 and BTV-6 vaccines, the level of variation observed (1.6%) suggests that this would have happened several cycles of replication before the virus arrived in northern Europe.
When BTV-6 was detected in the Netherlands during October 2008, it represented a further new serotype to Europe. Its African and vaccine strain origins are indicated by the phylogenetic analyses reported here. Recent studies also detected Seg-2 of the South African BTV-11 vaccine strain in the same part of northern Europe (in Belgium) during early 2009 [90]. Together with the arrival of BTV-8 in the Maastricht region during August 2006, this indicates that there is an effective route for the introduction of these viruses (none of which had previously been detected in Europe) to this part of northern Europe, although details of the mechanisms involved remain a matter for speculation.
The BTV-6 vaccine has previously been used are in Israel and South Africa itself. In each case it was used as part of a multivalent vaccine. In Israel this involved BTV-2, -4, -6, -10 and -16, while in South Africa the ‘Bottle-A’ vaccine contains BTV-1, -4, -6, -12 and -14. It has been suggested that the virus could have arrived in the region as the result of illegal use of these live attenuated BTV vaccines (‘Bottle-A’ of the south African BTV vaccine contains BTV-1, a strain of current concern in many regions of Europe). However, the absence of antibodies to multiple other serotypes in the antiserum from Heeten indicates that this animal at least had not received either of the multivalent vaccine preparations produced by Onderstepoort Biological Products (OBP) in South Africa.
Like most of the other genome segments (Seg-1 to 9), Seg-7/VP7 of NET2008/04 (KC isolate) and NET2008/05 (KC1/BHK1 isolate) showed 99.7-100% nt/aa identity to the South African BTV-6 live-vaccine strains. Similar results were also obtained with the virus after isolation in embryonated chicken eggs and passage in BHK-21 cells (at CVI). However, Seg-7 from the Heeten blood sample from (NET2008/06) generated a consensus Seg-7/VP7 sequence that was identical to Seg-7/VP7 of BTV-8 from the Netherlands 2006 (NET2006/04) and 2007 (NET2007/01), but only showed ∼80.5% nt identity to the BTV-6 isolates from the same sample (Figure 6). This suggests that the original blood contained two different versions of Seg-7 (consistent with the circulation of both BTV-6 and BTV-8 in the region), only one of which (from the tissue-culture adapted BTV-6 vaccine strain) was selected during isolation of the virus in cell-culture. It is possible that this reflects some involvement of the selected version of VP7 in the infection mechanism and consequently in adaptation of the virus to cell culture. However, the predominance of the BTV-8 Seg-7 in the original Heeten blood sample (NET2008/06) suggests that the virus was in the process of reassorting with the northern field strain of BTV-8 that was widespread in northern Europe.
We have considered the possibility that Seg-7 from the Netherlands strain of BTV-8 could have been detected in the blood sample from Heeten, as a result of laboratory contamination. However, our laboratory works to QA standards (ISO9001) and although the sequencing of Seg-7 was repeated three times, using freshly extracted RNA on each occasion, consistent results were obtained. Other samples extracted and tested at the same time did not contain BTV-8 Seg-7 and no other BTV-8 sequences (from other segments) were detected in the blood sample, collectively excluding the possibility of accidental contamination with BTV-8 Seg-7, or with BTV-8 virus.
In order to exchange genome segments these two ‘parental’ viruses (BTV-6 and BTV-8) must co-infect the same cells within an individual mammalian host or insect vector, and this appears most likely to have occurred after BTV-6 was introduced into northern Europe. Analyses of German 2008 samples by real-time RT-PCR (supplied by LSI) identified several animals that were infected by both BTV-6 and 8 (GER2008/01 and GER2008/02). These samples were also shown to contain a mix of the two different versions of Seg-7 detected from the Heeten blood and virus isolate.
It may be significant that evidence for reassortment was only detected in Seg7/VP7 and Seg-10/NS3/3a of the BTV-6 strain from northern Europe. VP7 is known to be involved in binding of BTV core particles to the cell surface membrane proteins of vector Culicoides sp and may play an important role in BTV infection of the insect vector [66], [91]. In contrast NS3/3a are involved in the release of virus particles from infected insect cells [67], [68], [92] and together with VP7 (and possibly the other outer coat protein VP2 and VP5 involved in cell attachment) may influence the dissemination of the virus within the individual insect. It has been suggested that variations in these two genome segments and proteins (Seg-7/VP7 and Seg10/NS3/3a) may reflect transmission of BTV by different vector species or populations in different geographic locations [35]. The acquisition of different versions of Seg-7 and Seg-10 by the vaccine strain of BTV-6 could therefore reflect adaptation to transmission by the vector population within the ecosystem that exists in northern Europe. However, further study will be needed to show if there are differences in the infection rate in north European Culicoides sp, or the transmission efficiency for these viruses containing the different versions of Seg-7/VP7.
The provision of a full genome sequence for the northern European field strain of BTV-6 will make it possible to track any further changes or reassortment events that occur if the virus continues to persist in the region.
Supporting Information
Accession numbers of BTV genome segments (Seg-7 and Seg-10) used in these phylogenetic analyses, in addition to those described previously by Maan et al [35], [57], [72].
(0.06 MB DOC)
Acknowledgments
The authors thank international colleagues who provided virus isolates for analyses and comparisons.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: SM and KN are funded by the Department for Environment, Food and Rural Affairs (Defra)(SE2617); NSM by the Biotechnology and Biological Sciences Research Council (BBSRC) and the European Union (EU) (SANCO/940/2002); PCM by Defra, the European Commission and BBSRC; PVR and RVG by the Ministry of Agriculture, Nature and Food Safety, The Netherlands; AS, CB and CO by Defra and the EU Commission; ACP and IW by the South African National Department of Agriculture, Forestry and Fisheries (DAFF); BH by the Federal Ministry of Food, Agriculture and Consumer Protection (Germany); ME by EPIZONE, the EU Network of Excellence for Epizootic Disease Diagnosis and Control (FOOD-CT-2006-016236). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mertens PPC, Maan S, Samuel A, Attoui H. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Orbivirus Reoviridae. . 2005. pp. 466–483. Virus Taxonomy VIIIth Report of the ICTV Elsevier/Academic Press London.
- 2.Fernandez-Pacheco P, Fernandez-Pinero J, Aguero M, Jiminez-Clavero MA. Bluetongue virus serotype 1 in wild mouflons in Spain. Vet Rec. 2008;162:659–660. doi: 10.1136/vr.162.20.659. [DOI] [PubMed] [Google Scholar]
- 3.Mauroy A, Guyot H, De Clercq K, Cassart D, Thiry E, et al. Bluetongue in captive yaks. Emerg Infect Dis. 2008;14:675–676. doi: 10.3201/eid1404.071416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barzilai E, Tadmor A. Multiplication of bluetongue virus in goats following experimental infection. Refu Vet. 1971;28:11–20. [Google Scholar]
- 5.Henrich M, Reinacher M, Hamann HP. Lethal bluetongue virus infection in an alpaca. Vet Rec. 2007;161:764. doi: 10.1136/vr.161.22.764. [DOI] [PubMed] [Google Scholar]
- 6.Meyer G, Lacroux C, Leger S, Top S, Goyeau K, et al. Lethal bluetongue virus serotype 1 infection in llamas. Emerg Infect Dis. 2009;15:608–610. doi: 10.3201/eid1504.081514. [DOI] [PubMed] [Google Scholar]
- 7.Ortega J, Crossley B, Dechant JE, Drew CP, Maclachlan NJ. Fatal bluetongue virus infection in an alpaca (Vicugna Pacos) in California. J Vet Diagn Invest. 2010;22:134–136. doi: 10.1177/104063871002200129. [DOI] [PubMed] [Google Scholar]
- 8.Darpel KE, Batten CA, Veronesi E, Williamson S, Anderson P, et al. Transplacental transmission of bluetongue virus 8 in cattle UK. Emerg Infect Dis. 2009;15:2025–2028. doi: 10.3201/eid1512.090788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Clerq K, Vandenbussche F, Vandemeulebroucke E, Vanbinst T, De Leeuw I, et al. Transplacental bluetongue infection in cattle. Vet Rec. 2008;162:564. doi: 10.1136/vr.162.17.564. [DOI] [PubMed] [Google Scholar]
- 10.Desmecht D, Bergh RV, Sartelet A, Leclerc M, Mignot C, et al. Evidence for transplacental transmission of the current wild-type strain of bluetongue virus serotype 8 in cattle. Vet Rec. 2008;163:50–52. doi: 10.1136/vr.163.2.50. [DOI] [PubMed] [Google Scholar]
- 11.Vercauteren G, Miry C, Vandenbussche F, Ducatelle R, Van der Heyden S, et al. Bluetongue virus serotype 8-associated congenital hydranencephaly in calves. Transbound Emerg Dis. 2008;55:293–298. doi: 10.1111/j.1865-1682.2008.01034.x. [DOI] [PubMed] [Google Scholar]
- 12.Wilson A, Darpel K, Mellor PS. Where does bluetongue virus sleep in the winter? PLoS Biol Aug. 2008;26; 6(8):e210. doi: 10.1371/journal.pbio.0060210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wouda W, Roumen MPHM, Peperkamp NHMT, Vos JH, van Garderen E, et al. Hydranencephaly in calves following the bluetongue serotype 8 epidemic in the Netherlands. Vet Rec. 2008;162:422–423. doi: 10.1136/vr.162.13.422-b. [DOI] [PubMed] [Google Scholar]
- 14.Backx A, Heutink R, van Rooij E, van Rijn P. Transplacental and oral transmission of wild-type bluetongue virus serotype 8 in cattle after experimental infection. Vet Microbiol. 2009;138:235–243. doi: 10.1016/j.vetmic.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Worwa G, Hilbe M, Ehrensperger F, Chaignat V, Hofmann MA, et al. Experimental transplacental bluetongue virus serotype 8 infection in sheep. Vet Rec. 2009;164:499–500. doi: 10.1136/vr.164.16.499. [DOI] [PubMed] [Google Scholar]
- 16.Alexander KA, MacLachlan NJ, Kat PW, House C, O'Brien SJ, et al. Evidence of natural bluetongue virus infection among African carnivores. Am J Trop Med Hyg. 1994;51:568–576. [PubMed] [Google Scholar]
- 17.Brown CC, Rhyan JC, Grubman MJ, Wilbur LA. Distribution of bluetongue virus in tissues of experimentally infected pregnant dogs as determined by in situ hybridization. Vet Pathol. 1996;33:337–340. doi: 10.1177/030098589603300311. [DOI] [PubMed] [Google Scholar]
- 18.Jauniaux TP, De Clercq KE, Cassart DE, Kennedy S, Vandenbussche FE, et al. Bluetongue in Eurasian lynx. Emerg Infect Dis. 2008;14:1496–1498. doi: 10.3201/eid1409.080434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 2006. 6th Ed, Chapter 219 Paris.
- 20.Darpel KE, Batten CA, Veronesi E, Shaw AE, Anthony S, et al. A study of British sheep and cattle infected with bluetongue virus serotype 8 from the 2006 outbreak in northern Europe. Vet Rec. 2007;161:253–261. doi: 10.1136/vr.161.8.253. [DOI] [PubMed] [Google Scholar]
- 21.Howell PG. Rome: FAO; 1963. Bluetongue Emerging Diseases of Animals; pp. 111–153. [Google Scholar]
- 22.Howell PG, Verwoerd DW. Bluetongue virus. In: Gard S, Hallauer C, Meyer KF, editors. Springer-Verlag. Vol 9. Vienna: Virology Monographs; 1971. pp. 35–74. [DOI] [PubMed] [Google Scholar]
- 23.Howerth EW, Greene CE, Prestwood AK. Experimentally induced bluetongue virus infection in white tailed deer: coagulation, clinical pathologic, and gross pathologic changes. Am J Vet Res. 1988;49:1906–1913. [PubMed] [Google Scholar]
- 24.Jeggo MJ, Corteyn AH, Taylor WP, Davidson WL, Gorman BM. Virulence of bluetongue virus for British sheep. Res Vet Sci. 1987;42:24–28. [PubMed] [Google Scholar]
- 25.MacLachlan NJ. The pathogenesis and immunology of bluetongue virus infection of ruminants. Comp Immunol Microbiol Infect Dis. 1994;17:197–206. doi: 10.1016/0147-9571(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 26.Vosdingh RA, Trainer DO, Easterday BC. Experimental bluetongue disease in white-tailed deer. Can J Comp Med Vet Sci. 1968;32:382–387. [PMC free article] [PubMed] [Google Scholar]
- 27.Callis J. Bluetongue in the United States. Prog Clin Biol Res. 1985;178:37–42. [PubMed] [Google Scholar]
- 28.Koumbati M, Mangana O, Nomikou K, Mellor PS, Papadopoulos O. Duration of bluetongue viraemia and serological responses in experimentally infected European breeds of sheep and goats. Vet Microbiol. 1999;64:277–285. doi: 10.1016/s0378-1135(98)00255-7. [DOI] [PubMed] [Google Scholar]
- 29.Parsonson IP. Pathology and pathogenesis of bluetongue infections. Curr Top Microbiol Immunol. 1990;162:119–141. doi: 10.1007/978-3-642-75247-6_5. [DOI] [PubMed] [Google Scholar]
- 30.Maclachlan NJ, Drew CP, Darpel KE, Worwa G. The Pathology and Pathogenesis of Bluetongue. J Comp Path. 2009;141:1–16. doi: 10.1016/j.jcpa.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Osburn BI. Bluetongue virus. Vet Clin North Am Food Anim Pract. 1994;10:547–560. doi: 10.1016/s0749-0720(15)30538-7. [DOI] [PubMed] [Google Scholar]
- 32.Hoogendam K. International study on the economic consequences of outbreaks of bluetongue serotype 8 in north-western Europe: Graduation Thesis University of Van Hall-Larenstein Leeuwarden 2007.
- 33.Velthuis AG, Saatkamp HW, Mourits MC, de Koeijer AA, Elbers AR. Financial consequences of the Dutch bluetongue serotype 8 epidemics of 2006 and 2007. Prev Vet Med. 2009;93:294–304. doi: 10.1016/j.prevetmed.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Mellor PS, Carpenter S, Harrup L, Baylis M, Wilson A, Mertens PPC. Bluetongue in Europe and the Mediterranean Basin. Chapter 13. In: Mellor PS, Baylis M, Mertens PPC, editors. Elsevier. London: Bluetongue; 2009. pp. 235–264. [Google Scholar]
- 35.Maan S, Maan NS, Ross-smith N, Batten CA, Shaw AE, et al. Sequence analysis of bluetongue virus serotype 8 from the Netherlands 2006 and comparison to other European strains. Virology. 2008;377:308–318. doi: 10.1016/j.virol.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 36.Mellor PS, Wittmann EJ. Bluetongue virus in the Mediterranean Basin 1998–2001. Vet J. 2002;164:20–37. doi: 10.1053/tvjl.2002.0713. [DOI] [PubMed] [Google Scholar]
- 37.Purse BV, Mellor PS, Rogers DJ, Samuel AR, Mertens PPC, et al. Climate change and the recent emergence of bluetongue in Europe. Nat Rev Microbiol 2: 171-181. Erratum in: Nat Rev Microbiol (2006) 2005;4:160. doi: 10.1038/nrmicro1090. [DOI] [PubMed] [Google Scholar]
- 38.Purse BV, Brown HE, Harrup L, Mertens PP, Rogers DJ. Invasion of bluetongue and other orbivirus infections into Europe: the role of biological and climatic processes. Rev Sci Tech. 2008;27:427–442. [PubMed] [Google Scholar]
- 39.Alpar OH, Bramwell WV, Veronesi E, Darpel EK, Pastore PP, Mertens PPC. Bluetongue virus vaccines past and present. Chapter 18. In: Mellor PS, Baylis M, Mertens PPC, editors. Elsevier. London: Bluetongue; 2009. pp. 235–264. [Google Scholar]
- 40.Dungu B, Potgieter C, Von Teichman B, Smit T. Vaccination in the control of bluetongue in endemic regions: the South African experience. Dev Biol (Basel) 2004;119:463–472. [PubMed] [Google Scholar]
- 41.Shimshony A. Bluetongue in Israel—a brief historical overview. Vet Ital. 2004;40:116–118. [PubMed] [Google Scholar]
- 42.Monaco F, Cammà C, Serini S, Savini G. Differentiation between field and vaccine strain of bluetongue virus serotype 16. Vet Microbiol. 2006;116:45–52. doi: 10.1016/j.vetmic.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 43.Listeš E, Monaco F, Labrović A, Paladini C, Leone A, et al. First evidence of bluetongue virus serotype 16 in Croatia. Vet Microbiol. 2009;138:92–97. doi: 10.1016/j.vetmic.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Savini G, MacLachlan NJ, Sanchez-Vizcaino JM, Zientara S. Vaccines against bluetongue in Europe. Comp Immunol Microbiol Infect Dis. 2008;31:101–120. doi: 10.1016/j.cimid.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Barros SC, Ramos F, Luis TM, Vaz A, Duarte M, et al. Molecular epidemiology of bluetongue virus in Portugal during 2004-2006 outbreak. Vet Microbiol. 2007;124:25–34. doi: 10.1016/j.vetmic.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 46.Batten CA, Maan S, Shaw AE, Maan NS, Mertens PPC. A European field strain of bluetongue virus derived from two parental vaccine strains by genome segment reassortment. Virus Res. 2008;137:56–63. doi: 10.1016/j.virusres.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Ferrari G, De Liberato C, Scavia G, Lorenzetti R, Zini M, et al. Active circulation of bluetongue vaccine virus serotype-2 among unvaccinated cattle in central Italy. Prev Vet Med. 2005;68:103–113. doi: 10.1016/j.prevetmed.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 48.Mellor PS, Carpenter S, Harrup L, Baylis M, Mertens PPC. Bluetongue in Europe and the Mediterranean Basin: history of occurrence prior to 2006. Prev Vet Med. 2008;87:4–20. doi: 10.1016/j.prevetmed.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Toussaint JF, Sailleau C, Mast J, Houdart P, Czaplicki G, et al. Bluetongue in Belgium 2006. Emerg Infect Dis. 2007;13:614–616. doi: 10.3201/eid1304.061136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verwoerd DW. Failure to demonstrate in vitro as opposed to in vivo transcription of the bluetongue virus genome. Onderstepoort J Vet Res. 1970;37:225–227. [PubMed] [Google Scholar]
- 51.Mertens PPC, Brown F, Sangar DV. Assignment of the genome segments of bluetongue virus type 1 to the proteins which they encode. Virology. 1984;135:207–217. doi: 10.1016/0042-6822(84)90131-4. [DOI] [PubMed] [Google Scholar]
- 52.Mertens PPC, Burroughs JN, Anderson J. Purification and properties of virus particles infectious sub-viral particles and cores of bluetongue virus serotypes 1 and 4. Virology. 1987a;157:375–386. doi: 10.1016/0042-6822(87)90280-7. [DOI] [PubMed] [Google Scholar]
- 53.Mertens PPC, Pedley S, Cowley J, Burrough JN. A comparison of six different bluetongue virus isolates by crosshybridization of the dsRNA genome segments. Virology. 1987b;161:438–447. doi: 10.1016/0042-6822(87)90137-1. [DOI] [PubMed] [Google Scholar]
- 54.Roy P. Bluetongue virus proteins. J Gen Virol. 1992;73:3051–3064. doi: 10.1099/0022-1317-73-12-3051. [DOI] [PubMed] [Google Scholar]
- 55.Verwoerd DW, Louw H, Oellermann RA. Characterization of bluetongue virus ribonucleic acid. J Virol. 1970;5:1–7. doi: 10.1128/jvi.5.1.1-7.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gould AR, Pritchard LI. Relationships amongst bluetongue viruses revealed by comparisons of capsid and outer coat protein nucleotide sequences. Virus Res. 1990;17:31–52. doi: 10.1016/0168-1702(90)90078-p. [DOI] [PubMed] [Google Scholar]
- 57.Maan S, Maan NS, Samuel AR, Rao S, Attoui H, et al. Analysis and phylogenetic comparisons of full-length VP2 genes of the 24 bluetongue virus serotypes. J Gen Virol. 2007a;88:621–630. doi: 10.1099/vir.0.82456-0. [DOI] [PubMed] [Google Scholar]
- 58.Pritchard LI, Gould AR, Wilson WC, Thompson L, Mertens PPC, et al. Complete nucleotide sequence of RNA segment 3 of bluetongue virus serotype 2 (Ona-A): Phylogenetic analyses reveal the probable origin and relationship with other orbiviruses. Virus Res. 1995;35:247–261. doi: 10.1016/0168-1702(94)00072-k. [DOI] [PubMed] [Google Scholar]
- 59.Pritchard LI, Sendow I, Lunt R, Hassan SH, Kattenbelt J, et al. Genetic diversity of bluetongue viruses in south east Asia. Virus Res. 2004;10:93–201. doi: 10.1016/j.virusres.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 60.Balasuriya UB, Nadler SA, Wilson WC, Pritchard LI, Smythe AB, et al. The NS3 proteins of global strains of bluetongue virus evolve into regional topotypes through negative (purifying) selection. Vet Microbiol. 2008;126:91–100. doi: 10.1016/j.vetmic.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 61.Nomikou K, Dovas CI, Maan S, Anthony SJ, Samuel AR, et al. Evolution and phylogenetic analysis of full-length VP3 genes of eastern Mediterranean bluetongue virus isolates. PLoS ONE. 2009;4(7):e6437. doi: 10.1371/journal.pone.0006437. doi: 101371/journalpone0006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cowley JA, Gorman BM. Cross neutralization of genetic reassortants of bluetongue virus serotypes 20 and 21. Vet Microbiol. 1989;19:37–51. doi: 10.1016/0378-1135(89)90089-8. [DOI] [PubMed] [Google Scholar]
- 63.Huismans H, Erasmus BJ. Identification of the serotype-specific and group specific antigens of bluetongue virus. Onderstepoort J Vet Res. 1981;48:51–58. [PubMed] [Google Scholar]
- 64.Mertens PPC, Pedley S, Cowley J, Burroughs JN, Corteyn AH, et al. Analysis of the roles of bluetongue virus outer capsid proteins VP2 and VP5 in determination of virus serotype. Virology. 1989;170:561–565. doi: 10.1016/0042-6822(89)90447-9. [DOI] [PubMed] [Google Scholar]
- 65.Hofmann MA, Renzullo S, Mader M, Chaignat V, Worwa G, et al. Genetic characterization of Toggenburg orbivirus a new bluetongue virus from goats Switzerland. Emerg Infect Dis. 2008;12:1855–1861. doi: 10.3201/eid1412.080818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mertens PPC, Burroughs JN, Walton A, Wellby MP, Fu H, et al. Enhanced infectivity of modified bluetongue virus particles for two insect cell lines and for two Culicoides vector species. Virology. 1996;217:582–593. doi: 10.1006/viro.1996.0153. [DOI] [PubMed] [Google Scholar]
- 67.Hyatt AD, Gould AR, Coupar B, Eaton BT. Localization of the non-structural protein NS3 in bluetongue virus-infected cells. J Gen Virol. 1991;72:2263–2267. doi: 10.1099/0022-1317-72-9-2263. [DOI] [PubMed] [Google Scholar]
- 68.Hyatt AD, Zhao Y, Roy P. Release of bluetongue virus-like particles from insect cells is mediated by BTV nonstructural protein NS3/NS3A. Virology. 1993;19:3592–3603. doi: 10.1006/viro.1993.1167. [DOI] [PubMed] [Google Scholar]
- 69.Bonneau KR, Zhang N, Zhu J, Zhang F, Li Z, et al. Sequence comparison of the L2 and S10 genes of bluetongue viruses from the United States and the People's Republic of China. Virus Res. 1999;61:153–160. doi: 10.1016/s0168-1702(99)00034-9. [DOI] [PubMed] [Google Scholar]
- 70.Bonneau KR, Zhang NZ, Wilson WC, Zhu JB, Zhang FQ, et al. Phylogenetic analysis of the S7 gene does not segregate Chinese strains of bluetongue virus into a single topotype. Arch Virol. 2000;145:1163–1171. doi: 10.1007/s007050070116. [DOI] [PubMed] [Google Scholar]
- 71.Wilson WC, Ma HC, Venter EH, van Djik AA, Seal BS, et al. Phylogenetic relationships of bluetongue viruses based on gene S7. Virus Res. 2000;67:141–151. doi: 10.1016/s0168-1702(00)00138-6. [DOI] [PubMed] [Google Scholar]
- 72.Maan S, Maan NS, Nomikou K, Anthony SJ, Ross-Smith N, et al. Molecular epidemiology studies of bluetongue virus Chapter 7. In: Mellor PS, Baylis M, Mertens PPC, editors. Elsevier. London: Bluetongue; 2009. pp. 135–156. [Google Scholar]
- 73.Vandenbussche F, De Leeuw I, Vandemeulebroucke E, De Clercq K. Emergence of bluetongue serotypes in Europe part 1: description and validation of four real-time RT-PCR assays for the serotyping of bluetongue viruses BTV-1 BTV-6 BTV-8 and BTV-11. Transbound Emerg Dis. 2009;56:346–354. doi: 10.1111/j.1865-1682.2009.01093.x. [DOI] [PubMed] [Google Scholar]
- 74.Eschbaumer M, Hoffmann B, Moss A, Savini G, Leone A, et al. Emergence of bluetongue virus serotype 6 in Europe—German field data and experimental infection of cattle. Vet Microbiol. 2009 doi: 10.1016/j.vetmic.2009.11.040. [Epub ahead of print] doi: 101016/jvetmic200911040. [DOI] [PubMed] [Google Scholar]
- 75.Grist NR, Ross CA, Bell EJ. Neutralization tests. Chapter 6. In: Grist NR, Ross CA, Bell EJ, editors. Blackwell Scientific Publications. Oxford London: Diagnostic Methods in Clinical Virology 2nd Edition; 1974. pp. 64–79. [Google Scholar]
- 76.Karber G. 50% end point calculation Archiv für Experimentelle Pathologie und Pharmakologie. 1931;162:480–483. [Google Scholar]
- 77.Attoui H, Billoir F, Cantaloube JF, Biagini P, de Micco P, et al. Strategies for the sequence determination of viral dsRNA genomes. J Virol Methods. 2000;89:147–158. doi: 10.1016/s0166-0934(00)00212-3. [DOI] [PubMed] [Google Scholar]
- 78.Maan S, Rao S, Maan NS, Anthony SJ, Attoui H, et al. Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J Virol Methods. 2007b;143:132–139. doi: 10.1016/j.jviromet.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 79.Shaw AE, Monaghan P, Alpar HO, Anthony S, Darpel KE, et al. Development and validation of a real-time RT-PCR assay to detect genome bluetongue virus segment 1. J Virol Methods. 2007;145:115–126. doi: 10.1016/j.jviromet.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 80.Mertens PPC, Maan NS, Prasad G, Samuel AR, Shaw AE, et al. Design of primers and use of RT-PCR assays for typing European bluetongue virus isolates: differentiation of field and vaccine strains. J Gen Virol. 2007;88:2811–2823. doi: 10.1099/vir.0.83023-0. [DOI] [PubMed] [Google Scholar]
- 81.Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, et al. Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol. 2009;90:1423–1432. doi: 10.1099/vir.0.009381-0. [DOI] [PubMed] [Google Scholar]
- 82.Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 40. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 84.Jeggo MH, Gumm ID, Taylor WP. Clinical and serological response of sheep to serial challenge with different bluetongue virus types. Res Vet Sci. 1983;34:205–211. [PubMed] [Google Scholar]
- 85.Jeggo MH, Wardley RC, Brownlie J, Corteyn AH. Serial inoculation of sheep with two bluetongue virus types. Res Vet Sci. 1986;40:386–392. [PubMed] [Google Scholar]
- 86.Wade-Evans AM, Mertens PPC, Belsham GJ. Sequence of genome segment 9 of bluetongue virus (serotype 1 South Africa) and expression analysis demonstrating that different forms of VP6 are derived from initiation of protein synthesis at two distinct sites. J Gen Virol. 1992;73:3023–3026. doi: 10.1099/0022-1317-73-11-3023. [DOI] [PubMed] [Google Scholar]
- 87.Mo CL, Thompson LH, Homan EJ, Oviedo MT, Greiner EC, et al. Bluetongue virus isolations from vectors and ruminants in Central America and the Caribbean Interamerican bluetongue team. Am J Vet Res. 1994;55:211–215. [PubMed] [Google Scholar]
- 88.Anthony SJ, Maan N, Maan S, Sutton GC, Attoui H, et al. Genetic and phylogenetic analysis of the core proteins VP1, VP3, VP4, VP6 and VP7 of epizootic haemorrhagic disease virus (EHDV). Virus Res. 2009;145:187–199. doi: 10.1016/j.virusres.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 89.Attoui H, Mohd Jaafar F, Belhouchet M, Aldrovandi N, Tao S, et al. Yunnan orbivirus, a new orbivirus species isolated from Culex tritaeniorhynchus mosquitoes in China. J Gen Virol. 2005;86:3409–3417. doi: 10.1099/vir.0.81258-0. [DOI] [PubMed] [Google Scholar]
- 90.De Clercq K, Mertens P, De Leeuw, Oura C, Houdart P, et al. Emergence of Bluetongue Serotypes in Europe Part 2: The Occurrence of a BTV-11 Strain in Belgium. Transbound Emerg Dis. 2009;56:355–361. doi: 10.1111/j.1865-1682.2009.01092.x. [DOI] [PubMed] [Google Scholar]
- 91.Xu G, Wilson W, Mecham J, Murphy K, Zhou EM, et al. VP7: an attachment protein of bluetongue virus for cellular receptors in Culicoides variipennis. . J Gen Virol. 1997;78:1617–1623. doi: 10.1099/0022-1317-78-7-1617. [DOI] [PubMed] [Google Scholar]
- 92.Hyatt AD, Eaton BT, Brookes SM. The release of bluetongue virus from infected cells and their superinfection by progeny virus. Virology. 1989;173:21–34. doi: 10.1016/0042-6822(89)90218-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession numbers of BTV genome segments (Seg-7 and Seg-10) used in these phylogenetic analyses, in addition to those described previously by Maan et al [35], [57], [72].
(0.06 MB DOC)