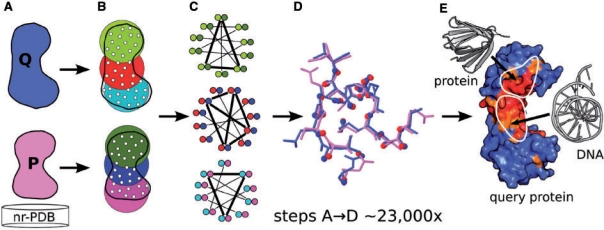
Fig. 2.
Schematic representation of the ProBiS algorithm. (A) The query protein structure (Q) is compared in a pairwise manner with each of ∼23 000 non-redundant structures (P). (B) Proteins, represented as graphs of vertices (white dots) and edges (not shown), are divided into n overlapping subgraphs, where n equals the number of vertices and all vertices are within 15 Å of a central vertex: three subgraphs per protein are depicted here as distinctly colored encirclements. A fast distance-matrix-based filtering is applied to them to eliminate non-similar subgraphs. (C) A product graph is constructed for each similar pair of subgraphs (see color encoding in B and C). A maximum clique (thick lines) in a product graph represents the largest similarity between two compared protein subgraphs. (D) Each maximum clique produces a structural alignment of two compared proteins (the alignment shown corresponds to the middle maximum clique in C). (E) Steps A–D are repeated for each protein from the nr-PDB and the results are stored in a MySQL database. Structural similarity scores are calculated and projected on the query protein surface. Structurally similar and variable residues are colored red and blue, respectively. High-scoring residues are considered as predicted structurally similar binding sites.