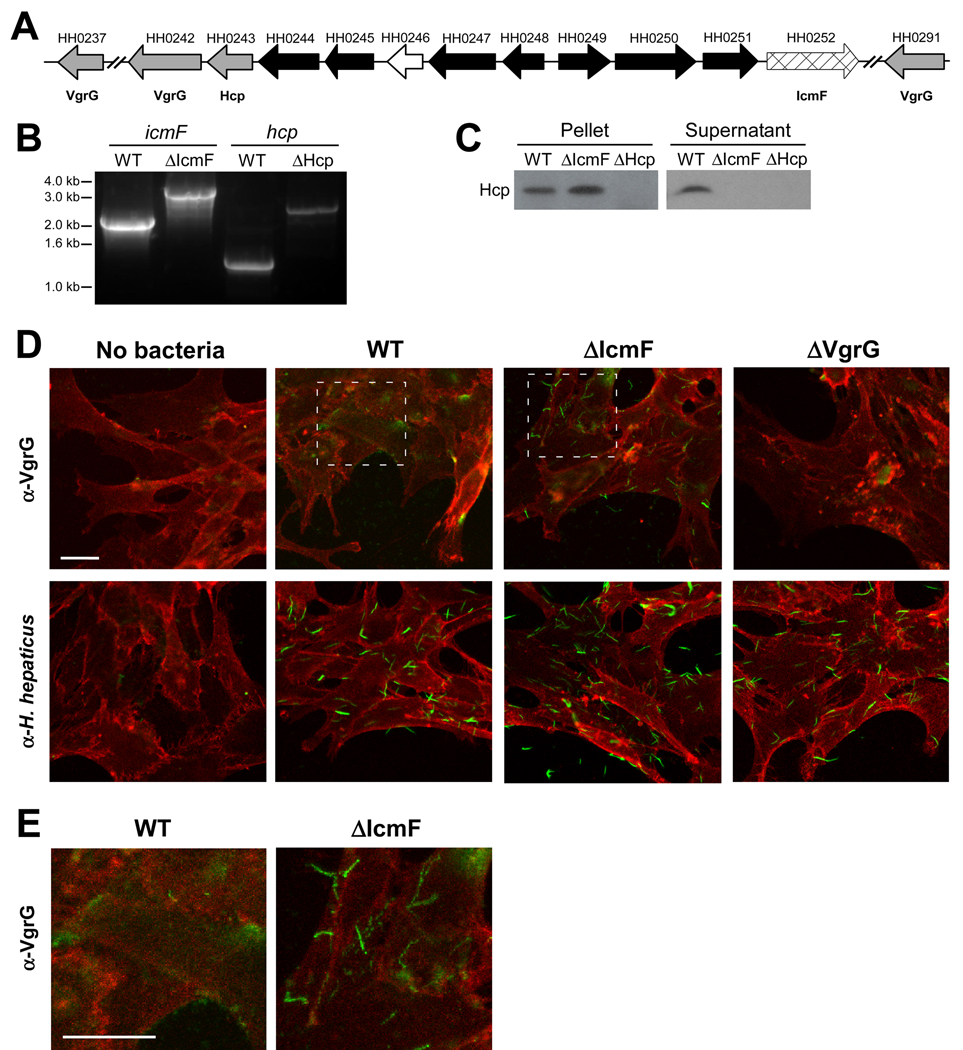
Figure 1. H. hepaticus Encodes for a Functional T6SS.
(A) Schematic diagram of the genetic organization of H. hepaticus T6SS genes. Gray arrows represent Hcp and VgrG genes, cross-hatched arrow indicates IcmF homologue, black arrows represent T6SS homologues of unknown function, and white arrow indicates a gene non-homologous to other T6SS genes. See also Table S1.
(B) Genomic DNA collected from mid-log cultures of WT, ΔIcmF, or ΔHcp H. hepaticus was amplified by PCR using icmF or hcp specific primers. Insertion of the eryR gene was detected by a 1.1kb-increase in the resulting PCR band.
(C) Hcp is undetected in supernatants from ΔIcmF and ΔHcp T6SS mutants. Equal amounts of mid-log bacterial cultures of WT, ΔIcmF, or ΔHcp were centrifuged to separate bacterial pellets and supernatant. Supernatants were subsequently filtered to ensure removal of all bacteria. Bacterial pellets and supernatants were analyzed by Western blot. Membranes were blotted with anti-Hcp antibody.
(D, E) Confocal images of bacteria incubated with MODE-K cells. WT, ΔIcmF, or ΔVgrG (ΔHH0242) H. hepaticus were incubated with MODE-K cells for 5hr. MODE-K cells were subsequently rinsed with PBS, fixed in 4% PFA, and stained for the eukaryotic cell membrane marker wheat germ agglutinin (red) and either H. hepaticus or VgrG (green). Outlined regions for WT and ΔIcmF in (D) are shown at higher magnification (E). Scale bar represents 20µm.