Fig. 5.
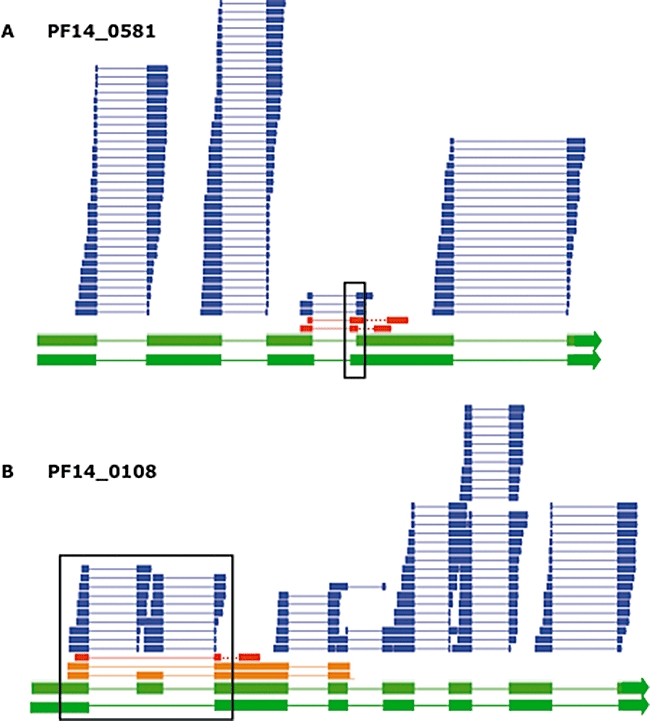
Use of RNA-Seq data to detect alternative splicing and exon skipping events in the IDC transcriptome of P. falciparum 3D7. A. Alternative splice sites for exon 4 of PF14_0581 (putative apicoplast ribosomal protein isoforms) highlighted by aligned bridging reads (red) from early ring time points. The boxed area highlights the location of alternative splicing in the gene PF14_0581. The dotted red line links read pairs from the same template DNA. The blue bars show reads that map to the borders of exons, across an intron. Perfectly mapping reads are not shown. B. Example of exon skipping in PF14_0108 (a predicted protein of unknown function). A new splice form was indicated by a read mapping across two introns and exon, and its read pair (red). Both splice-forms were confirmed by RT-PCR (orange boxes). The boxed area highlights the location of exon skipping in the gene PF14_0108. Only the last eight exons of PF14_0108 are shown in the figure.
