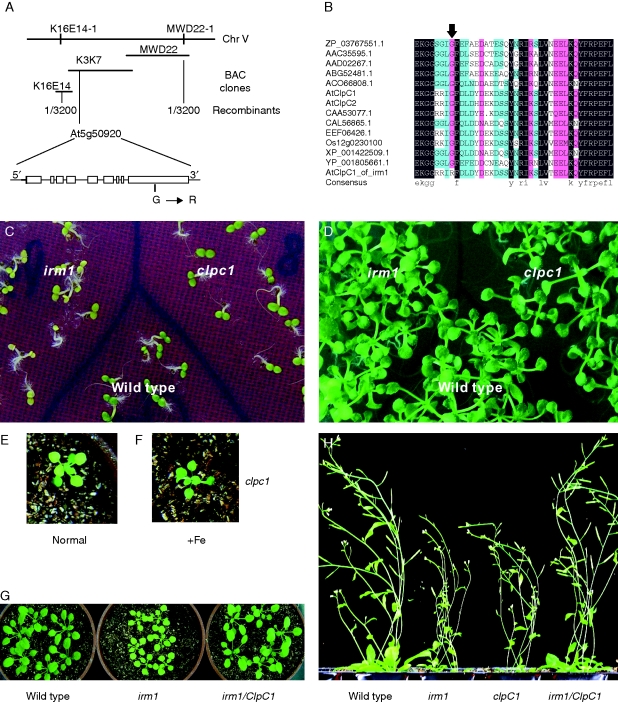
Fig. 4.
(A) The schematic outline of genetic and physical maps of IRM1 and the structure of the candidate gene At5g50920 (ClpC1) localized on BAC clone K3K7 (the open boxes represent exons and the lines among the open boxes represent introns). The site of mutation G → R is shown. (B) Sequence alignment of ClpC proteins from Arabidopsis (AtClpC1 and AtClpC2), Nostoc azollae (ZP_03767551·1), Trichodesmium erythraeum (ABG52481·1), Guillardia theta (AAC35595·1), Rhodomonas salina (AAD02267·1), Micromonas sp. RCC299 (AC066808·1), Populus trichocarpa (EEF06426·1), Brassica napus (CAA53077·1), Ostreococcus tauri (CAL56865·1), Oryza sativa (Os12g0230100), Ostreococcus lucimarinus CCE9901 (XP_001422509·1) and Cyanothece sp. ATCC 51142 (YP_001805661·1). The amino acid indicated by arrow is the mutation site found in the irm1 mutant. (C and D) Phenotypic comparison of irm1, clpc1 (a T-DNA insertion mutant of ClpC1) and wild type grown on an MS agar plate for 4 d (C) and for 10 d (D). (E and F) Phenotypic recovery of leaf chlorosis of clpc1 in soil, 3 d after being watered with Fe solution (100 µm FeEDTA) (+Fe). (G and H) Phenotypic complementation by expression of ClpC1 in an irm1 mutant at early (G) and a late stage (H).