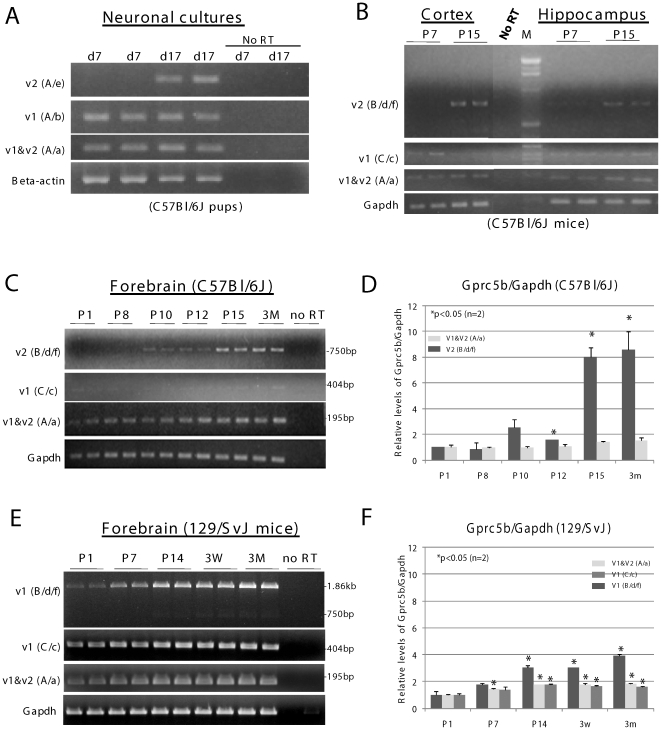
Figure 4. Expression of Gprc5b_v2 is increased in maturing neurons and during postnatal brain development.
A. cDNA prepared from C57Bl/6J cortical neurons cultured for 7 days (d7) or 17 days (d17) was amplified with primer sets A/e, A/b, and A/a to detect Gprc5b_v2, Gprc5b_v1, and a region common to both splice variants, respectively. Cortical neurons were isolated from C57Bl/6J pups at postnatal day 0 (P0). Beta-actin mRNA was amplified as a control. B. Expressions of Gprc5b_v2, and Gprc5b_v1 in cortex and hippocampus of C57Bl/6J postnatal day 7 (P7) and day 15 (P15). Primer sets B/d/f, C/c, and A/a were used in RT-PCR to detect Gprc5b_v2, Gprc5b_v1, and a region common to both splice variants, respectively. C. Expressions of Gprc5b_v2, and Gprc5b_v1 in forebrains of C57Bl/6J mice during brain development. cDNA prepared from C57Bl/6J forebrains, was amplified with the same primer sets described in B. The tissues were collected from mouse pups at postnatal days 1 through 15 (P1–P15) and from 3 month-old (3M) adult mice. D. Quantitative analysis of expressions of Gprc5b splice variants detected in C. Relative intensities of Gprc5b_v2 (top panel of C), v1 & v2 combined (the 3rd panel from top of C), and Gapdh (bottom panel of C) cDNA bands were determined by densitometry analysis with ScionImage software (www.scioncorp.com). Levels of Gprc5b were normalized to those of Gapdh. The two-tailed t-test was used for the statistical analysis (all comparisons were made to P1). Levels of Gprc5b_v1 (the 2nd panel from top of C) were not quantified due to its low expression. E. Expressions of Gprc5b_v1, and Gprc5b_v2 in forebrains of 129/SvJ mice during brain development. cDNA prepared from 129/SvJ forebrains were amplified with the same primer sets described in B. The tissues were collected from mouse pups at postnatal days 1 through 14 (P1–P14) and at three weeks (3W), and from 3 month-old (3M) adult mice. F. Quantitative analysis of expressions of Gprc5b splice variants detected in E, as described in D. See Fig. 1A for relative positions of primers used. For B–F, Gapdh mRNA was amplified as a control. No RT: mRNA that was not reverse transcribed to make cDNA. M: DNA markers.