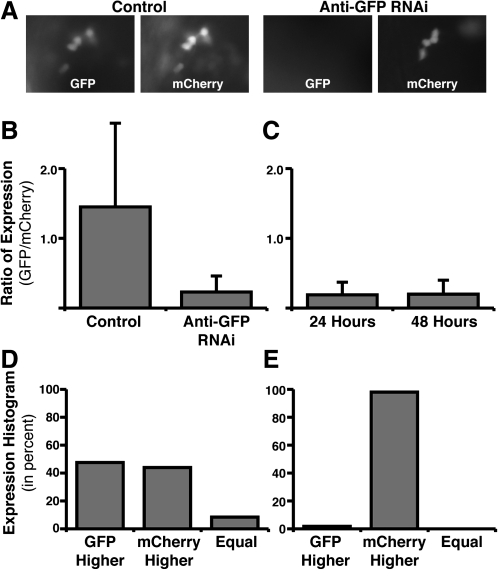
FIG. 6.
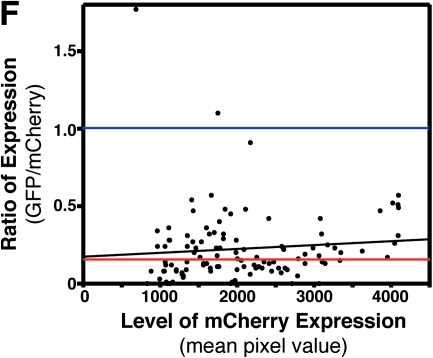
Using in vivo electroporation for RNA interference (RNAi)–based loss of function. (A) Fluorescent images showing that coelectroporation of an anti-GFP-targeted siRNA along with an expression plasmid for GFP leads to decreased GFP fluorescence as compared to cells electroporated with a nontargeting siRNA. A plasmid coding for mCherry was also included to identify the cells successfully electroporated. (B) The degree to which GFP expression was blocked was quantified by ratioing GFP to mCherry fluorescence in single cells (see Materials and Methods). Control electroporations included a nontargeting siRNA, a GFP expression plasmid, and an mCherry expression plasmid. Anti-GFP RNAi electroporations included an siRNA targeting GFP, a GFP expression plasmid, and an mCherry expression plasmid. (C) The degree to which GFP expression was blocked was analyzed in the same embryos 24 and 48 h after electroporation. (D) Histogram of all control cells analyzed. The cells have been categorized by the GFP/mCherry ratio for three different categories: (i) GFP higher expressed, (ii) mCherry higher expressed, or (iii) equal expression of GFP and mCherry (within 5%). (E) Histogram for all anti-GFP RNAi cells analyzed. (F) The GFP/mCherry ratio for all analyzed anti-GFP RNAi cells plotted against the total mCherry fluorescence (mean pixel value). This graph represents the degree of knockdown (ratio) as a function of total expression via the plasmids (mCherry expression). Black line represents a linear regression of all RNAi data. Red line represents the median ratio for RNAi cells. Blue line represents the median ratio for all control cells (note: control cells are not plotted on the graph).