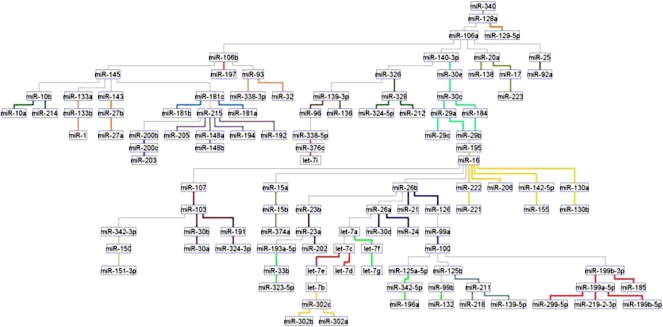
Figure 1.
The miRNA network in normal tissues (1107 samples, 50 tissues, 115 miRNAs). The network was inferred for all expressed and varying miRNAs, without preselecting for differential expression. Standard Banjo parameters were adopted with a q6 discretization policy. The consensus graph depicted here was obtained from the best 100 nets (after searching through 8.3 × 109 networks). MCL graph-based clustering algorithm was applied to clusters extraction (miRNAs with highly related expression pattern have edges of the same color); thus, different clusters in the network are linked by different color edges. yEd graph editor (yFiles software) was employed for graphs visualization. See text for further discussion.