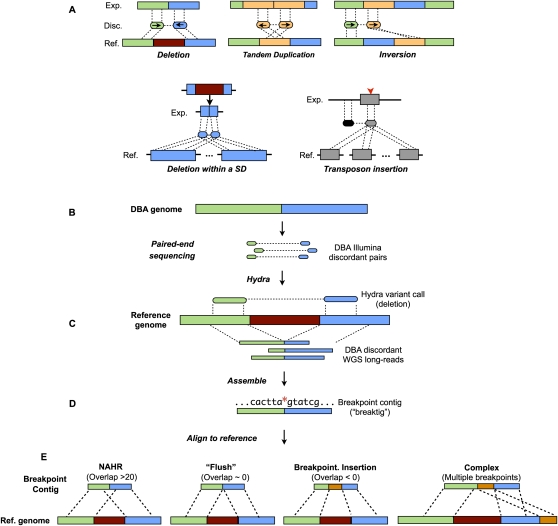
Figure 1.
Overview of structural variation discovery pipeline. (A) Paired-end mapping signatures are shown for five different classes of structural variation as detected by paired-end mapping. Notably, each end of a given discordant matepair will map to each copy of a segmental duplication (blue, bottom left panel) when a mutation arises in one copy. In the case of a new transposon insertion (gray rectangle with red arrowhead, bottom right panel), ends of discordant matepairs that originated from the newly inserted sequence will map to all other similar elements in the genome. (Exp.) Experimental genome; (Disc.) discordant pairs from experimental genome; (Ref.) reference genome; (SD) segmental duplication. (B) Matepairs from the DBA strain are aligned to the mouse reference genome. (C) Clusters of discordant matepairs (often with multiple possible mapping combinations) are identified by HYDRA as putative variants (a deletion is shown). (D) Discordant WGS long-reads that corroborate the HYDRA call are assembled into breakpoint contigs (“breaktigs”) with phrap. The red asterisk indicates the nucleotide at which the SV breakpoint occurred. (E) Breaktigs are then aligned to the reference genome with MEGABLAST using very sensitive settings. The observed sequence homology (evident as alignment “overlap” at the breakpoint) in the resulting alignments is a hallmark of the causal SV mechanism, where negative overlap indicates the presence of an insertion or small-scale rearrangements directly at the breakpoint. (NAHR) Non-allelic homologous recombination.