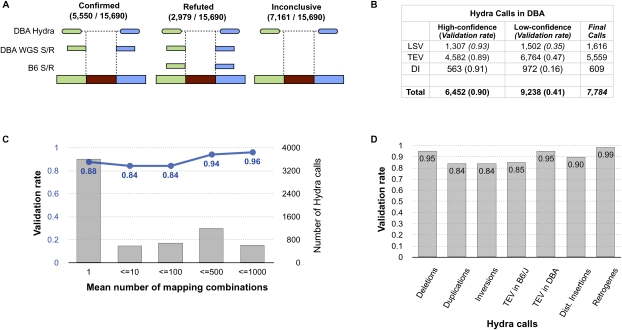
Figure 2.
Validation of HYDRA calls. (A) HYDRA breakpoint calls in DBA were compared to split-read (S/R) alignments of WGS long-reads from both DBA and B6. Calls in DBA are corroborated by split-read mapping(s) from DBA that map within the predicted breakpoint interval in an orientation that is consistent with the HYDRA call. However, if one or more split-reads from B6 supports the call, it is refuted under the assumption that it originated from a mapping artifact or an error in the reference genome assembly. Cases in which split-reads were observed in neither strain were deemed inconclusive. Due to the relatively low coverage of DBA WGS long-reads, many HYDRA calls were inconclusive. (B) The number and validation rate for high-confidence and low-confidence HYDRA calls are shown for DBA. The 7784 final HYDRA calls represent the high-confidence calls that were not refuted by split-reads plus the low-confidence calls that were confirmed. (LSV) Local SVs, such as duplications, deletions, and inversions; (TEV) transposable element variants; (DI) “distant” insertions of non-transposon DNA from >1 Mb away (including retrogenes). For a detailed table describing the different SV classes and their validation rates, see Supplemental Table S1. (C) The validation rate (blue) and number (gray) of HYDRA SV calls is shown as a function of the mean number of mapping combinations observed among the supporting matepairs. (D) A comparison of the validation rate of HYDRA SV calls by the type of variant.