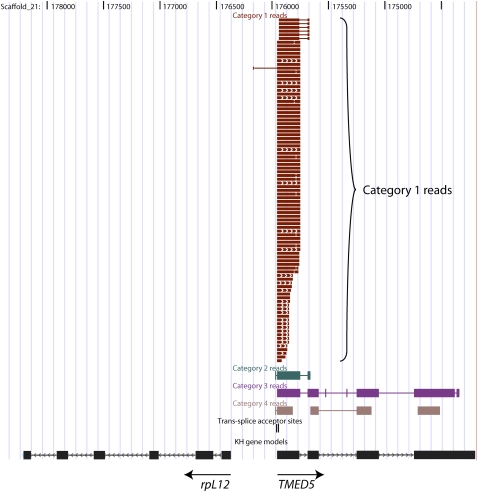
Figure 2.
Example of SL-PCR reads mapping to a trans-spliced gene. The figure shows a ∼4-kb region of the Ciona genome (assembly version 1), as viewed in the UCSC Genome Browser (http://genome.ucsc.edu), edited for clarity, with SL-PCR reads mapped to the genome by BLAT. In addition to Category 1 reads, i.e., reads containing a perfect SL primer motif at one end or the other, other read categories shown as separate tracks are Category 2 (imperfect SL primer motif at one end or the other), Category 3 (AN-start reads terminating within the SL-PCR product insert and thus falling short of the SL primer at the far end), and Category 4 (atypical reads that do not start with a recognizable primer motif, or that contain additional unexpected primer motifs within the SL-PCR insert). The Category 1 read track is displayed in “squish” mode and the Categories 2–4 tracks in “dense” mode. Also shown is a track indicating with vertical strokes the locations of 8929 trans-splice acceptor sites mapped to the genome on the basis of Category 1 reads, two of which map close together at the 5′-end the TMED5 (transmembrane emp24 protein transport domain containing 5) gene.