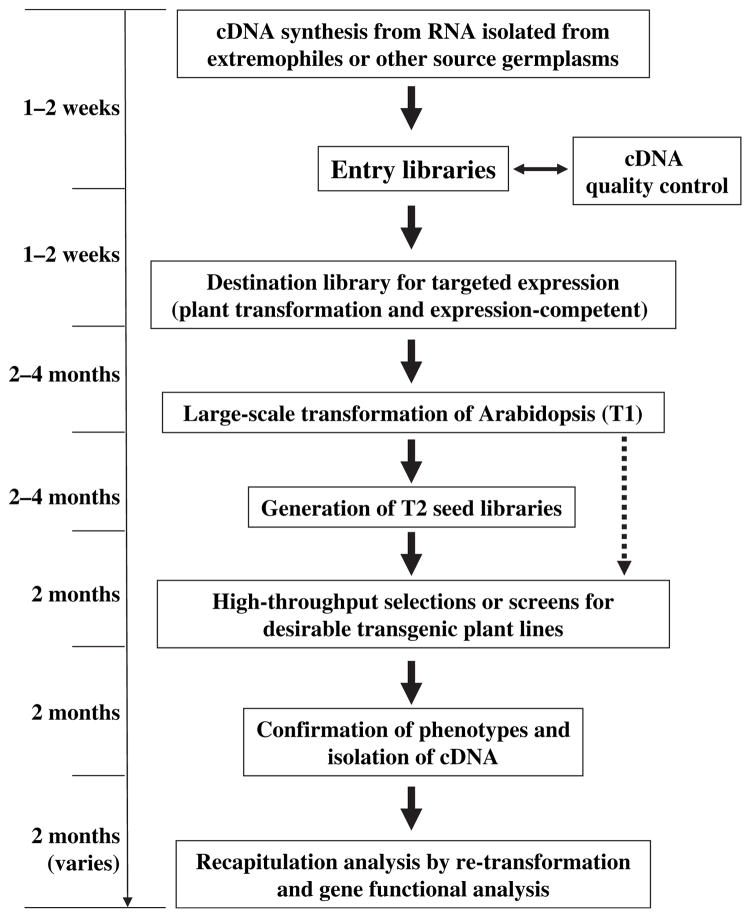
Figure 1.
Flow chart illustrating the functional gene-mining method with the power of Arabidopsis, and the minimal time required for each step.
Starting from cDNA synthesis for the source germplasms (for example, salt cress), through entry and destination expression library construction, large-scale transformation of Arabidopsis to generate the T2 seed library, primary high-throughput screen of the T2 seed library (T1 seeds can also be screened if the T2 library is not maintained) for salt tolerance, for example, secondary screen to confirm the phenotype and isolation of cDNA, to the final recapitulation and gene function analysis: the whole procedure requires a minimal time of 1 year. The broken arrow indicates an alternative route.