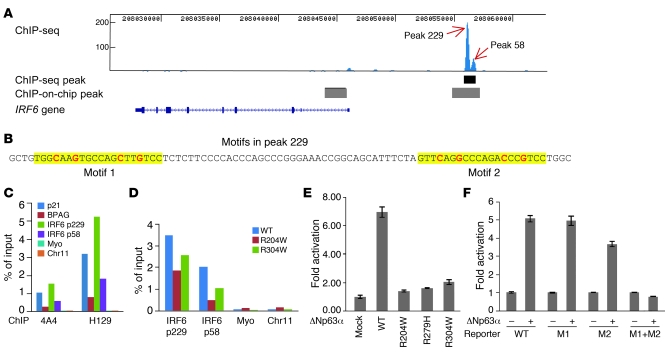
Figure 5. A p63 binding site upstream of IRF6 functions as an enhancer element.
(A) ChIP-seq analysis of p63 binding in human primary keratinocytes. The p63 binding site obtained from ChIP-seq is shown as a black bar below the double peak (peak 229 and peak 58), and p63 binding sites reported previously by ChIP-on-chip (19) are shown as gray bars. (B) The 2 p63 binding motifs identified within peak 229 are highlighted in yellow, with the most conserved cytosine and guanine bases shown in red. (C) ChIP-qPCR analysis of p63 binding using p63 antibodies 4A4 and H129. Specific binding of p63 to the positive controls p21 and BPAG as well as to peak 229 (p229) and peak 58 (p58) regions, but not to negative controls myoglobin exon 2 (myo) and a no-gene region (chr11), was observed. (D) ChIP-qPCR analysis of R204W and R304W cell lines indicated reduced p63 binding. (E) Transient transfection assays showed that wild-type p63 strongly activated transcription through peak 229. In contrast, activation by the p63 mutants R204W, R279H, and R304W was greatly reduced, with the highest level (approximately 2-fold) in R304W. (F) Site-directed mutagenesis of the conserved cytosine and guanine bases showed that both motifs in peak 229 were responsive to p63 and that mutation abolished transactivation. WT, reporter of wild-type peak 229; M1, mutation of motif 1; M2, mutation of motif 2; M1+M2, mutation of both motifs.